Molecularly Imprinted Polymer-Based Biomimetic Systems for Sensing Environmental Contaminants, Biomarkers, and Bioimaging Applications
Abstract
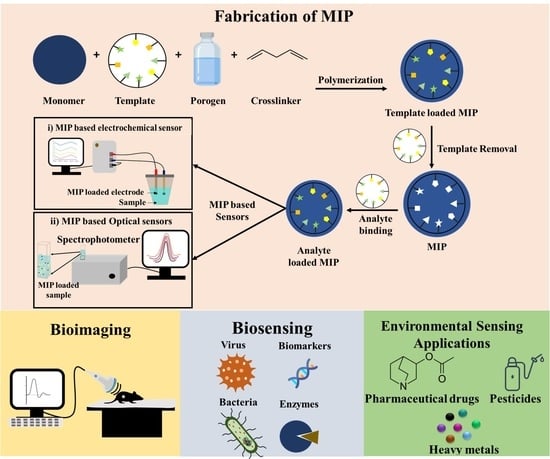
:1. Introduction
2. Preparation Methods for MIPs
2.1. Bulk Polymerization
2.2. Suspension Polymerization
2.3. Emulsion Polymerization
2.4. Precipitation Polymerization
2.5. Multi-Step Swelling Polymerization
2.6. Surface Imprinting Polymerization
2.7. Electrochemical Polymerization
3. Imprinting Techniques for MIPs
3.1. Covalent Imprinting
3.2. Non-Covalent Imprinting
3.3. Semi-Covalent Interactions
3.4. Metal-Mediated Interactions
4. MIPs-Based Sensors
4.1. MIP-Based Optical Sensors
4.1.1. MIPs-Based Optical Sensors for Pharmaceutical Drug Detection
4.1.2. MIPs in the Detection of Bacteria and Viruses
4.1.3. MIPs in Bioimaging
5. MIP-Based Electrochemical Sensor
5.1. MIP-Based Electrochemical Sensor in Environmental Applications
| Synthesis Method | Functional Monomer | Detection Method | Analyte | LoD | Recovery Real Sample | Reference |
|---|---|---|---|---|---|---|
| Precipitation polymerization | Vinyl benzene, MAA | DPV | Chloridazon | 6.2 × 10−8 mol L−1 | Ground water-95% Surface water-94% Drinking water-96.5% Sea water-92% | [136] |
| Precipitation polymerization | 2-vinylpyridine, AM, MAA | DPAdCSV | Hexazinone | 2.6 × 10−12 mol L−1 | River water-95.8% | [137] |
| Precipitation polymerization | MAA, 2-(5-Bromo-2-pyridylazo)-5-(diethylamino)phenol | DPAdCSV | Uranyl Ions | 1.1 × 10−10 mol L−1 | Tap water-99.8% Caspian Sea water-100.4% Persian Gulf water-100.7% River water-99.5% | [138] |
| Methylene succinic acid | Potentiometric | Cr (III) | 5.9 × 10−7 mol L−1 | River water-98% Sea water-102% | [139] | |
| Precipitation polymerization | MAA | Voltammetric | Para-nitrophenol | 3 × 10−9 mol L−1 | Tap water-99.4% River water-100.4% | [140] |
| MAA | Square wave voltammetry | Dicloran | 9.4 × 10−10 mol L−1 | Tap water-96.50% River water-100.30% Urine-93% | [141] | |
| Core-shell | MAA | Square wave voltammetry | TNT | 0.5 nM | Tap water-(94–100.6%) Sea water-(90–107.5%) | [142] |
| MAA, 4-aminothiophenol | CV and DPV | Tetrabromobisphenol-S | 0.029 nM | Tap water-(98.7–107.3%) | [143] | |
| Bulk polymerization | 2-vinylpyridine | SWV | Diuron | 9.0 × 10−9 mol L−1 | River water-(96.1–99.5%) | [144] |
| Precipitation polymerization | Vinyl pyridine, MAA | SWV | cerium (III) | 10 pM | Drinking water-(95–97.3%) Sea water-(102.7–10.4%) | [145] |
| Bulk polymerization | MAA | SWV | Carbofuran | 3 × 10–10 M | Tap water-(94–96%) River water-(94–97%) Urine-(91–94%) | [146] |
| Radical polymerization | MAA | DPV | Diphenylamine | 0.1 mM | Synthetic sample | [147] |
| Electro polymerization | DPV | Cd2+ | 1.62 × 10−4 μm | Tap water-(98.5–102.2%) River water-(99.5–100.67%) Milk-(99–109.2%) | [148] | |
| Sol-gel | 3-[2-(2-aminoethylamino) ethylamino] propyl-trimethoxysilane | DPASV | Cd2+ | 0.15 μgL−1 | Tap water and River water- (97.0–101.7%) | [149] |
| Suspension Polymerization | MAA | DPV | Methylene blue | 11.65 µg/mL | - | [150] |
| Precipitation polymerization | MAA | SWV | Paraoxon | 1.0 × 10−9 mol L−1 | Tap water-(101.3%) River water-(103.2%) Lake water-(97.8%) | [151] |
| Sol gel | 3-Aminopropyl triethoxysilane | DPV | Tetrabromobisphenol-A | 0.77 nM | Tap water-(96.54–105.78%) Pool water-(92.41–99.27%) | [130] |
| Precipitation polymerization | MAA | DPV | Pb2+ | 1.3 × 10−11 mol L−1 | Flour-(99.1%) Rice-(103.7%) Tap water-(99.4%) River water-(102.1%) | [152] |
| Precipitation polymerization | MAA Ethylene glycol dimethacrylate | potentiometric sensor | Cu2+ | 2.0 × 10−9 mol L−1 | Tap water-(101–103%) River water-(100–106%) | [153] |
| Precipitation polymerization | MAA | DPV | Ag(I) | 97 μg L−1 | Well water-(97.2–98.2%) Aqueduct water-(98.2–103%) Dam water-(97.3–99.6%) | [154] |
| Precipitation polymerization | MAA | Impedimetric sensor | 5-Chloro-2,4-dinitrotoluene | 0.1 μM | [155] | |
| Precipitation polymerization | AM | square-wave adsorptive anodic strippingvoltammetry | Methyl green | 1.0 × 10−8 mol L−1 | River water-(99.5–103%) Industrial waste water-(93.7–99.3%) | [156] |
| Electro polymerization | ortho-phenylenediamine | DPV | Acesulfame-K | 0.35 µM | Cool drink-(100.8–108%) Candy-(99.6–104%) Tabletop sweetener-(98.4–102.4) | [157] |
| Electro polymerization | pyrrole | DPV | catechol | 0.54 µM | Tap water-(93.90 to 99.69% | [158] |
| Electro polymerization | l-arginine | Cyclic voltammetry | Tartrazine | 0.0027 µM | Soft water-(92.63–105.59%) Orange-flavored jelly powder-(95–100.7%) | [159] |
| Bulk polymerization | Itaconic acid | DPV | Metribuzin | 0.1 pg/mL | Pure samples-(99.29–101.38%) Tomatoes samples-(98.74–102.34%) Potatoes sample-(97.47–103.4%) | [160] |
| Electro polymerization | o-phenylenediamine | DPV | Nitrofurazone | 0.18 nmol L−1 | Milk-(96.06–101.46%) | [161] |
| Electro polymerization | MAA | DPV | ceftriaxone | 0.008 µM | Powder-(98.67–101.71%) Urine-(101.44–104.20%) | [162] |
| MAA | DPV | creatinine | 5.9 × 10−8 M | Plasma samples-(97.40–119.25%) | [163] | |
| Electro polymerization | o-Phenylenediamine | DPV | Thiabendazole | 0.23 μM | Apple-(78.2–86.4%) Pear-(87.7–91.2%) Orange juice-(82.3–87.1) | [164] |
| Electro polymerization | Pyrrole | DPV | picric acid | 1.4 μM | - | [165] |
| Radical polymerization | MAA | DPV | maleic hydrazide | 40 ppb | Onion-(88.5–94.5%) Garlic-(82.2–105.1%) Potato-(80.0–106%) | [166] |
| Thermal precipitation polymerization | MAA | Voltammetric | 2,4-dinitrophenol and 2,4,6 trinitrotoluene | 0.59 μM and 0.29 μM | [167] | |
| Electro polymerization | Carbazole | SWV | Nitrobenzene | 0.107 μM | Honey-(99–114%) | [168] |
| Thermal polymerization | MAA; itaconic acid; acrylamide; 2-(trifluoromethyl)-acrylic acid; N, N-Methylene Bis Acrylamide | EIS | Methidathion | 5.14 μg/L | Tap water-(98–100.35) | [110] |
| Precipitation polymerization | MAA | EIS | N-nitrosodimethylamine | 0.85 μg/L | Tap water-(99%) | [169] |
| Self-polymerization | Dopamine | EIS | Dichlorodiphenyltrichloroethane | 6 × 10−12 mol L−1 | Raddish-(83–102%) | [170] |
| Electropolymerization | o-Phenylenediamine | EIS | Alachlor | 0.8 nM | Tap water-(95.5–103.5%) | [171] |
5.2. MIPs-Based Electrochemical Sensors for Bio Applications
| Synthesis Method | Functional Monomer | Detection Method | Analyte | LOD | Recovery Real Sample | Reference |
|---|---|---|---|---|---|---|
| Electro polymerization | 3-aminophenol | Amperometry | Tau-441 protein | 0.01 pmol/L | [181] | |
| Electro polymerization | Methylene blue | DPV | Lysosome | 4.26 fM | Serum-(94–108%) Urea-(98–109%) | [182] |
| Electro polymerization | DPV | Immunoglobulin G | 0.017 ngmL−1 | Serum-(97.36–100.98%) | [110] | |
| Electro polymerization | Aniline | CV and EIS | Histamine | 1.07 nM | - | [183] |
| Electro polymerization | polyacrylamide | DPV | Dopamine andadenine | 0.12–0.37 μM and 0.15–0.37 μM | Serum-dopamine (96–108%) Serum-adenine (92–104%) | [164] |
| Chemical polymerization | Aniline | EIS | Tryptophan | 8 pM | Milk-(98.4–101.4%) | [184] |
| Electro polymerization | DPV | Cortisol | 20.2 pM | [185] | ||
| Electro polymerization | poly o-phenylenediamine | CV, EIS, and SWV | Cortisol | 200 fM | Saliva-(91–105%) | [186] |
| Electro polymerization | CV and EIS | Aflatoxin B1 | 12.0 pg L−1 | Milk-(97–104%) | [187] | |
| Photopolymerization | MAA | DPV | Cholesterol and cholestanol | 0.01 μM | Serum-(93.6–101.03%) | [188] |
| Photopolymerization | AM | EIS | Neutrophil gelatinase-associated lipocalin (NGAL) | 0.07μg/mL | Real NGAL-91% | [189] |
| Electro polymerization | EIS | SARS-CoV-2 | 10 to 108 PFU/mL | Saliva-(98 to 104%) | [190] | |
| Free radical polymerization | vinyl phosphonic acid | sarcosine | 0.04 µM | [191] | ||
| One pot method | DPV | Creatinine | 2 × 10−2 pg/mL | Serum, urine (93.7–109.2%) | [192] | |
| Methyl methacrylate | DPV | H. pylori | 0.05 ng mL−1 | Blood-96% | [193] | |
| Electro polymerization | 2-aminophenol | EIS | Galectin-3 | 30 ng/mL | [194] | |
| Electro polymerization | Dopamine | DPV | Trypsin | 0.75 pg/mL | Urine-(94–100.2%) Serum-(98.4–114%) | [195] |
| Precipitation polymerization | Methyl methacrylate | DPV | serum amyloid A | 0.01 pM | [196] | |
| Electro polymerization | Methyl methacrylate | EIS | Follicle-stimulating hormone (FSH) | 0.1 pM | Blood samples (90–98.79) | [197] |
| Electro polymerization | Eriochrome Black T | EIS | Interleukin-1β | 1.5 pM | [198] | |
| Co-Electropolymerization | carboxylated pyrrole | EIS | Interleukin-6 (IL-6) | 0.02 pg/mL | [199] | |
| Bulk polymerization | Methyl methacrylate | CV and EIS | Anandamide | 0.01 nM | Blood samples-(93.48 and 90.08%) | [200] |
| Electro polymerization | 3-aminophenylboronic acid | DPV | Lactate | 0.22 μM | Sugarcane vinasse-(97.7 to 104.8%) | [109] |
| Electro polymerization | 3-aminophenylboronic acid | CV and EIS | Interleukin-6 | 1 pg/mL | [179] |
6. Conclusions and Future Perspectives
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Berglund, L.; Björling, E.; Oksvold, P.; Fagerberg, L.; Asplund, A.; Szigyarto, C.A.-K.; Persson, A.; Ottosson, J.; Wernérus, H.; Nilsson, P.; et al. A Genecentric Human Protein Atlas for Expression Profiles Based on Antibodies. Mol. Cell. Proteom. 2008, 7, 2019–2027. [Google Scholar] [CrossRef] [Green Version]
- Gray, A.C.; Bradbury, A.R.M.; Knappik, A.; Plückthun, A.; Borrebaeck, C.A.K.; Dübel, S. Animal-derived-antibody generation faces strict reform in accordance with European Union policy on animal use. Nat. Methods 2020, 17, 755–756. [Google Scholar] [CrossRef] [PubMed]
- Tchekwagep, P.M.S.; Crapnell, R.D.; Banks, C.E.; Betlem, K.; Rinner, U.; Canfarotta, F.; Lowdon, J.W.; Eersels, K.; van Grinsven, B.; Peeters, M.; et al. A Critical Review on the Use of Molecular Imprinting for Trace Heavy Metal and Micropollutant Detection. Chemosensors 2022, 10, 296. [Google Scholar] [CrossRef]
- Satheeshkumar, P.K. Expression of Single Chain Variable Fragment (scFv) Molecules in Plants: A Comprehensive Update. Mol. Biotechnol. 2020, 62, 151–167. [Google Scholar] [CrossRef] [Green Version]
- Crivianu-Gaita, V.; Thompson, M. Aptamers, antibody scFv, and antibody Fab’ fragments: An overview and comparison of three of the most versatile biosensor biorecognition elements. Biosens. Bioelectron. 2016, 85, 32–45. [Google Scholar] [CrossRef] [PubMed]
- Jayasena, S.D. Aptamers: An emerging class of molecules that rival antibodies in diagnostics. Clin. Chem. 1999, 45, 1628–1650. [Google Scholar] [CrossRef] [Green Version]
- Crapnell, R.D.; Hudson, A.; Foster, C.W.; Eersels, K.; van Grinsven, B.; Cleij, T.J.; Banks, C.E.; Peeters, M. Recent Advances in Electrosynthesized Molecularly Imprinted Polymer Sensing Platforms for Bioanalyte Detection. Sensors 2019, 19, 1204. [Google Scholar] [CrossRef] [Green Version]
- Hasseb, A.A.; Ghani, N.D.T.A.; Shehab, O.R.; El Nashar, R.M. Application of molecularly imprinted polymers for electrochemical detection of some important biomedical markers and pathogens. Curr. Opin. Electrochem. 2022, 31, 100848. [Google Scholar] [CrossRef]
- Vasapollo, G.; Del Sole, R.; Mergola, L.; Lazzoi, M.R.; Scardino, A.; Scorrano, S.; Mele, G.; Vasapollo, G.; Del Sole, R.; Mergola, L.; et al. Molecularly Imprinted Polymers: Present and Future Prospective. Int. J. Mol. Sci. 2011, 12, 5908–5945. [Google Scholar] [CrossRef] [Green Version]
- Asman, S.; Mohamad, S.; Sarih, N.M. Exploiting β-Cyclodextrin in Molecular Imprinting for Achieving Recognition of Benzylparaben in Aqueous Media. Int. J. Mol. Sci. 2015, 16, 3656–3676. [Google Scholar] [CrossRef] [Green Version]
- Pardo, A.; Mespouille, L.; Dubois, P.; Blankert, B.; Duez, P. Molecularly Imprinted Polymers: Compromise between Flexibility and Rigidity for Improving Capture of Template Analogues. Chem.—A Eur. J. 2014, 20, 3500–3509. [Google Scholar] [CrossRef] [PubMed]
- Wulff, G.; Grobe-Einsler, R.; Vesper, W.D.; Sarhan, A.A. Enzyme-analogue built polymers, 5. On the specificity dis-tribution of chiral cavities prepared in synthetic polymers†. Macromol. Chem. Phys. 1977, 178, 2817–2825. [Google Scholar] [CrossRef]
- Whitcombe, M.J.; Rodriguez, M.E.; Villar, P.; Vulfson, E.N. A New Method for the Introduction of Recognition Site Functionality into Polymers Prepared by Molecular Imprinting: Synthesis and Characterization of Polymeric Receptors for Cholesterol. J. Am. Chem. Soc. 1995, 117, 7105–7111. [Google Scholar] [CrossRef]
- Arshady, R.; Mosbach, K. Synthesis of substrate-selective polymers by host-guest polymerization. Die Makromol. Chem. 1981, 182, 687–692. [Google Scholar] [CrossRef]
- Öter, Ç.; Zorer, Ö.S. Molecularly imprinted polymer synthesis and selective solid phase extraction applications for the detection of ziram, a dithiocarbamate fungicide. Chem. Eng. J. Adv. 2021, 7, 100118. [Google Scholar] [CrossRef]
- Wang, G.N.; Yang, K.; Liu, H.Z.; Feng, M.X.; Wang, J.P. Molecularly imprinted polymer-based solid phase extraction combined high performance liquid chromatography for determination of fluoroquinolones in milk. Anal. Methods 2016, 8, 5511–5518. [Google Scholar] [CrossRef]
- Yang, S.; Wang, Y.; Jiang, Y.; Li, S.; Liu, W. Molecularly Imprinted Polymers for the Identification and Separation of Chiral Drugs and Biomolecules. Polymers 2016, 8, 216. [Google Scholar] [CrossRef]
- Wei, W.; Zhou, T.; Wu, S.; Shen, X.; Zhu, M.; Li, S. An enzyme-like imprinted-polymer reactor with segregated quantum confinements for a tandem catalyst. RSC Adv. 2018, 8, 1610–1620. [Google Scholar] [CrossRef] [Green Version]
- Kurczewska, J.; Cegłowski, M.; Pecyna, P.; Ratajczak, M.; Gajęcka, M.; Schroeder, G. Molecularly imprinted polymer as drug delivery carrier in alginate dressing. Mater. Lett. 2017, 201, 46–49. [Google Scholar] [CrossRef]
- Lantigua, D.; Nguyen, M.A.; Wu, X.; Suvarnapathaki, S.; Kwon, S.; Gavin, W.; Camci-Unal, G. Synthesis and characterization of photocrosslinkable albumin-based hydrogels for biomedical applications. Soft Matter 2020, 16, 9242–9252. [Google Scholar] [CrossRef]
- Zheng, W.; Wu, H.; Jiang, Y.; Xu, J.; Li, X.; Zhang, W.; Qiu, F. A molecularly-imprinted-electrochemical-sensor modified with nano-carbon-dots with high sensitivity and selectivity for rapid determination of glucose. Anal. Biochem. 2018, 555, 42–49. [Google Scholar] [CrossRef]
- De León-Martínez, L.D.; Rodríguez-Aguilar, M.; Ocampo-Pérez, R.; Gutiérrez-Hernández, J.M.; Díaz-Barriga, F.; Batres-Esquivel, L.; Flores-Ramírez, R. Synthesis and Evaluation of a Molecularly Imprinted Polymer for the Determination of Metronidazole in Water Samples. Bull. Environ. Contam. Toxicol. 2018, 100, 395–401. [Google Scholar] [CrossRef]
- Ghasemi, S.; Nematollahzadeh, A. Molecularly imprinted ultrafiltration polysulfone membrane with specific nano-cavities for selective separation and enrichment of paclitaxel from plant extract. React. Funct. Polym. 2018, 126, 9–19. [Google Scholar] [CrossRef]
- BelBruno, J.J. Molecularly Imprinted Polymers. Chem. Rev. 2019, 119, 94–119. [Google Scholar] [CrossRef] [PubMed]
- Mustafa, Y.L.; Keirouz, A.; Leese, H.S. Molecularly imprinted polymers in diagnostics: Accessing analytes in biofluids. J. Mater. Chem. B 2022, 10, 7418–7449. [Google Scholar] [CrossRef]
- Sharma, G.; Kandasubramanian, B. Molecularly Imprinted Polymers for Selective Recognition and Extraction of Heavy Metal Ions and Toxic Dyes. J. Chem. Eng. Data 2020, 65, 396–418. [Google Scholar] [CrossRef]
- Fu, J.; Wang, X.; Li, J.; Ding, Y.; Chen, L. Synthesis of multi-ion imprinted polymers based on dithizone chelation for simultaneous removal of Hg2+, Cd2+, Ni2+ and Cu2+ from aqueous solutions. RSC Adv. 2016, 6, 44087–44095. [Google Scholar] [CrossRef] [Green Version]
- Liu, W.; Qin, L.; An, Z.; Chen, L.; Liu, X.; Yang, Y.; Xu, B. Thermo-responsive ion imprinted polymer on the surface of magnetic carbon microspheres for identification and removal of low-concentrations of Cu2+. Environ. Chem. 2018, 15, 306. [Google Scholar] [CrossRef]
- Rahangdale, D.; Kumar, A.; Archana, G.; Dhodapkar, R.S. Ion cum molecularly dual imprinted polymer for simultaneous removal of cadmium and salicylic acid. J. Mol. Recognit. 2018, 31, e2630. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, L.; Zhu, F.; Lu, Y.; Guan, J. Synthesis, adsorption and selectivity of inverse emulsion Cd(II) imprinted polymers. Chin. J. Chem. Eng. 2018, 26, 494–500. [Google Scholar] [CrossRef]
- Jalilzadeh, M.; Uzun, L.; Şenel, S.; Denizli, A. Specific heavy metal ion recovery with ion-imprinted cryogels. J. Appl. Polym. Sci. 2016, 133. [Google Scholar] [CrossRef]
- Tamahkar, E.; Bakhshpour, M.; Andaç, M.; Denizli, A. Ion imprinted cryogels for selective removal of Ni(II) ions from aqueous solutions. Sep. Purif. Technol. 2017, 179, 36–44. [Google Scholar] [CrossRef]
- Karnka, R.; Chaiyasat, P.; Chaiyasat, A. Synthesis of Uniform and Stable Molecularly Imprinted Polymer Particles by Precipitation Polymerization. Orient. J. Chem. 2017, 33, 2370–2376. [Google Scholar] [CrossRef]
- Li, Y.; Song, H.; Zhang, L.; Zuo, P.; Ye, B.-C.; Yao, J.; Chen, W. Supportless electrochemical sensor based on molecularly imprinted polymer modified nanoporous microrod for determination of dopamine at trace level. Biosens. Bioelectron. 2016, 78, 308–314. [Google Scholar] [CrossRef] [PubMed]
- Eddin, F.B.K.; Fen, Y.W. Recent Advances in Electrochemical and Optical Sensing of Dopamine. Sensors 2020, 20, 1039. [Google Scholar] [CrossRef] [Green Version]
- Kamarudin, S.F.; Ahmad, M.N.; Dzahir, I.H.M.; Ishak, N.; Ab Halim, N.F. Development of quercetin imprinted membranes-based PVDF substrate. Polym. Bull. 2019, 76, 4313–4334. [Google Scholar] [CrossRef]
- Patel, K.D.; Kim, H.; Knowles, J.C.; Poma, A. Molecularly Imprinted Polymers and Electrospinning: Manufacturing Convergence for Next-Level Applications. Adv. Funct. Mater. 2020, 30, 2001955. [Google Scholar] [CrossRef]
- Wang, J.; Cormack, P.A.G.; Sherrington, D.C.; Khoshdel, E. Monodisperse, Molecularly Imprinted Polymer Microspheres Prepared by Precipitation Polymerization for Affinity Separation Applications. Angew. Chem. Int. Ed. 2003, 42, 5336–5338. [Google Scholar] [CrossRef] [PubMed]
- Rong, F.; Feng, X.; Li, P.; Yuan, C.; Fu, D. Preparation of molecularly imprinted microspheres by photo-grafting on supports modified with iniferter. Chin. Sci. Bull. 2006, 51, 2566–2571. [Google Scholar] [CrossRef]
- Yang, J.; Li, Y.; Wang, J.; Sun, X.; Cao, R.; Sun, H.; Huang, C.; Chen, J. Molecularly imprinted polymer microspheres prepared by Pickering emulsion polymerization for selective solid-phase extraction of eight bisphenols from human urine samples. Anal. Chim. Acta 2015, 872, 35–45. [Google Scholar] [CrossRef]
- Ho, K.-C.; Yeh, W.-M.; Tung, T.-S.; Liao, J.-Y. Amperometric detection of morphine based on poly(3,4-ethylenedioxythiophene) immobilized molecularly imprinted polymer particles prepared by precipitation polymerization. Anal. Chim. Acta 2005, 542, 90–96. [Google Scholar] [CrossRef]
- Lai, J.-P.; Yang, M.-L.; Niessner, R.; Knopp, D. Molecularly imprinted microspheres and nanospheres for di(2-ethylhexyl)phthalate prepared by precipitation polymerization. Anal. Bioanal. Chem. 2007, 389, 405–412. [Google Scholar] [CrossRef]
- Kou, X.; Lei, J.; Geng, L.; Deng, H.; Jiang, Q.; Zhang, G.; Ma, G.; Su, Z. Synthesis, characterization and adsorption behavior of molecularly imprinted nanospheres for erythromycin using precipitation polymerization. J. Nanosci. Nanotechnol. 2012, 12, 7388–7394. [Google Scholar] [CrossRef]
- He, C.; Ledezma, U.H.; Gurnani, P.; Albelha, T.; Thurecht, K.J.; Correia, R.; Morgan, S.; Patel, P.; Alexander, C.; Korposh, S. Surface polymer imprinted optical fibre sensor for dose detection of dabrafenib. Analyst 2020, 145, 4504–4511. [Google Scholar] [CrossRef]
- Lu, H.; Tian, H.; Wang, C.; Xu, S. Designing and controlling the morphology of spherical molecularly imprinted polymers. Mater. Adv. 2020, 1, 2182–2201. [Google Scholar] [CrossRef]
- Tian, Y.; Wang, Y.; Wu, S.; Sun, Z.; Gong, B. Preparation of Ampicillin Surface Molecularly Imprinted Polymers for Its Selective Recognition of Ampicillin in Eggs Samples. Int. J. Anal. Chem. 2018, 2018, 5897381. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Xu, S.; Li, J. Recent advances in molecular imprinting technology: Current status, challenges and highlighted applications. Chem. Soc. Rev. 2011, 40, 2922–2942. [Google Scholar] [CrossRef] [PubMed]
- Sambe, H.; Hoshina, K.; Haginaka, J. Molecularly imprinted polymers for triazine herbicides prepared by multi-step swelling and polymerization method: Their application to the determination of methylthiotriazine herbicides in river water. J. Chromatogr. A 2007, 1152, 130–137. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, Y.; Masumoto, S.; Kubo, A.; Matsunaga, H.; Haginaka, J. Preparation of molecularly imprinted polymers for warfarin and coumachlor by multi-step swelling and polymerization method and their imprinting effects. J. Chromatogr. A 2017, 1516, 71–78. [Google Scholar] [CrossRef]
- Haginaka, J.; Sanbe, H. Uniformly sized molecularly imprinted polymer for (S)-naproxen: Retention and molecular recognition properties in aqueous mobile phase. J. Chromatogr. A 2001, 913, 141–146. [Google Scholar] [CrossRef]
- Haginaka, J. Synthesis of Molecularly Imprinted Polymers by Two-Step Swelling and Polymerization; Humana: New York, NY, USA, 2021; Volume 2359, pp. 1–8. [Google Scholar] [CrossRef]
- Wegner, G.; Wernet, W.; Glatzhofer, D.; Ulanski, J.; Kröhnke, C.; Mohammadi, M. Chemistry and conductivity of some salts of polypyrrole. Synth. Met. 1987, 18, 1–6. [Google Scholar] [CrossRef]
- Peng, H.; Yin, F.; Zhou, A.; Yao, S. Characterization of electrosynthesized poly- (o-aminophenol) as a molecular imprinting material for sensor preparation by means of quartz crystal impedance analysis. Anal. Lett. 2002, 35, 435–450. [Google Scholar] [CrossRef]
- Yongabi, D.; Khorshid, M.; Losada-Pérez, P.; Eersels, K.; Deschaume, O.; D’Haen, J.; Bartic, C.; Hooyberghs, J.; Thoelen, R.; Wübbenhorst, M.; et al. Cell detection by surface imprinted polymers SIPs: A study to unravel the recognition mechanisms. Sensors Actuators B Chem. 2018, 255, 907–917. [Google Scholar] [CrossRef]
- Murdaya, N.; Triadenda, A.L.; Rahayu, D.; Hasanah, A.N. A Review: Using Multiple Templates for Molecular Imprinted Polymer: Is It Good? Polymers 2022, 14, 4441. [Google Scholar] [CrossRef]
- Mujahid, A.; Iqbal, N.; Afzal, A. Bioimprinting strategies: From soft lithography to biomimetic sensors and beyond. Biotechnol. Adv. 2013, 31, 1435–1447. [Google Scholar] [CrossRef]
- Panasyuk, T.; Orto, V.C.D.; Marrazza, G.; El’Skaya, A.; Piletsky, S.; Rezzano, I.; Mascini, M. Molecular Imprinted Polymers Prepared by Electropolymerization of Ni-(Protoporphyrin IX). Anal. Lett. 1998, 31, 1809–1824. [Google Scholar] [CrossRef]
- Poma, A.; Turner, A.P.; Piletsky, S.A. Advances in the manufacture of MIP nanoparticles. Trends Biotechnol. 2010, 28, 629–637. [Google Scholar] [CrossRef] [PubMed]
- Mayes, A.G.; Mosbach, K. Molecularly Imprinted Polymer Beads: Suspension Polymerization Using a Liquid Perfluorocarbon as the Dispersing Phase. Anal. Chem. 1996, 68, 3769–3774. [Google Scholar] [CrossRef]
- Adumitrăchioaie, A. Electrochemical Methods Based on Molecularly Imprinted Polymers for Drug Detection. A Review. Int. J. Electrochem. Sci. 2018, 13, 2556–2576. [Google Scholar] [CrossRef]
- Wulff, G.; Wolf, G. Zur Chemie von Haftgruppen, VI. Über die Eignung verschiedener Aldehyde und Ketone als Haftgruppen für Monoalkohole. Eur. J. Inorg. Chem. 1986, 119, 1876–1889. [Google Scholar] [CrossRef]
- Zhang, L.; Xu, J.S.; Sanders, V.M.; Letson, A.D.; Roberts, C.J.; Xu, R.X. Multifunctional microbubbles for image-guided antivascular endothelial growth factor therapy. J. Biomed. Opt. 2010, 15, 030515. [Google Scholar] [CrossRef]
- Komaba, S.; Seyama, M.; Momma, T.; Osaka, T. Potentiometric biosensor for urea based on electropolymerized electroinactive polypyrrole. Electrochimica Acta 1997, 42, 383–388. [Google Scholar] [CrossRef]
- Shea, K.J.; Sasaki, D.Y. An analysis of small-molecule binding to functionalized synthetic polymers by 13C CP/MAS NMR and FT-IR spectroscopy. J. Am. Chem. Soc. 1991, 113, 4109–4120. [Google Scholar] [CrossRef]
- Sallacan, N.; Zayats, M.; Bourenko, T.; Kharitonov, A.B.; Willner, I. Imprinting of Nucleotide and Monosaccharide Recognition Sites in Acrylamidephenylboronic Acid−Acrylamide Copolymer Membranes Associated with Electronic Transducers. Anal. Chem. 2002, 74, 702–712. [Google Scholar] [CrossRef]
- Wulff, G.; Vietmeier, J. Enzyme-analogue built polymers, 26. Enantioselective synthesis of amino acids using polymers possessing chiral cavities obtained by an imprinting procedure with template molecules. Macromol. Chem. Phys. 1989, 190, 1727–1735. [Google Scholar] [CrossRef]
- Yu, L.; Sun, L.; Zhang, Q.; Zhou, Y.; Zhang, J.; Yang, B.; Xu, B.; Xu, Q. Nanomaterials-Based Ion-Imprinted Electrochemical Sensors for Heavy Metal Ions Detection: A Review. Biosensors 2022, 12, 1096. [Google Scholar] [CrossRef] [PubMed]
- Yan, H.; Row, K.H. Characteristic and Synthetic Approach of Molecularly Imprinted Polymer. Int. J. Mol. Sci. 2006, 7, 155–178. [Google Scholar] [CrossRef] [Green Version]
- Wulff, G. Enzyme-like Catalysis by Molecularly Imprinted Polymers. Chem. Rev. 2002, 102, 1–28. [Google Scholar] [CrossRef]
- Sellergren, B.; Lepistoe, M.; Mosbach, K. Highly enantioselective and substrate-selective polymers obtained by molecular imprinting utilizing noncovalent interactions. NMR and chromatographic studies on the nature of recognition. J. Am. Chem. Soc. 1988, 110, 5853–5860. [Google Scholar] [CrossRef]
- Dickert, F.L.; Tortschanoff, M.; Bulst, W.E.; Fischerauer, G. Molecularly Imprinted Sensor Layers for the Detection of Polycyclic Aromatic Hydrocarbons in Water. Anal. Chem. 1999, 71, 4559–4563. [Google Scholar] [CrossRef]
- Sellergren, B.; Andersson, L. Molecular recognition in macroporous polymers prepared by a substrate analog imprinting strategy. J. Org. Chem. 1990, 55, 3381–3383. [Google Scholar] [CrossRef]
- Joshi, V.; Karode, S.; Kulkarni, M.; Mashelkar, R. Novel separation strategies based on molecularly imprinted adsorbents. Chem. Eng. Sci. 1998, 53, 2271–2284. [Google Scholar] [CrossRef]
- Alexander, C.; Andersson, H.; Andersson, L.I.; Ansell, R.J.; Kirsch, N.; Nicholls, I.A.; O’Mahony, J.; Whitcombe, M.J. Molecular imprinting science and technology: A survey of the literature for the years up to and including 2003. J. Mol. Recognit. 2006, 19, 106–180. [Google Scholar] [CrossRef] [PubMed]
- Kyzas, G.Z.; Bikiaris, D.N. Molecular Imprinting for High-Added Value Metals: An Overview of Recent Environmental Applications. Adv. Mater. Sci. Eng. 2014, 2014, 932637. [Google Scholar] [CrossRef] [Green Version]
- Cacho, C.; Schweitz, L.; Turiel, E.; Pérez-Conde, C. Molecularly imprinted capillary electrochromatography for selective determination of thiabendazole in citrus samples. J. Chromatogr. A 2008, 1179, 216–223. [Google Scholar] [CrossRef]
- Fang, L.; Liao, X.; Jia, B.; Shi, L.; Kang, L.; Zhou, L.; Kong, W. Recent progress in immunosensors for pesticides. Biosens. Bioelectron. 2020, 164, 112255. [Google Scholar] [CrossRef] [PubMed]
- Ye, T.; Yin, W.; Zhu, N.; Yuan, M.; Cao, H.; Yu, J.; Gou, Z.; Wang, X.; Zhu, H.; Reyihanguli, A.; et al. Colorimetric detection of pyrethroid metabolite by using surface molecularly imprinted polymer. Sensors Actuators B Chem. 2018, 254, 417–423. [Google Scholar] [CrossRef]
- Xiao, Y.; Qian, X. Substitution of oxygen with silicon: A big step forward for fluorescent dyes in life science. Coord. Chem. Rev. 2020, 423, 213513. [Google Scholar] [CrossRef]
- Qu, Y.; Qian, H.; Mi, Y.; He, J.; Gao, H.; Lu, R.; Zhang, S.; Zhou, W. Rapid determination of the pesticide ametryn based on a colorimetric aptasensor of gold nanoparticles. Anal. Methods 2020, 12, 1919–1925. [Google Scholar] [CrossRef]
- Sergeyeva, T.; Yarynka, D.; Piletska, E.; Linnik, R.; Zaporozhets, O.; Brovko, O.; Piletsky, S.; El’Skaya, A. Development of a smartphone-based biomimetic sensor for aflatoxin B1 detection using molecularly imprinted polymer membranes. Talanta 2019, 201, 204–210. [Google Scholar] [CrossRef]
- Singh, A.; Dhiman, N.; Kar, A.K.; Singh, D.; Purohit, M.P.; Ghosh, D.; Patnaik, S. Advances in controlled release pesticide formulations: Prospects to safer integrated pest management and sustainable agriculture. J. Hazard. Mater. 2020, 385, 121525. [Google Scholar] [CrossRef] [PubMed]
- Huang, C.; Hu, B. Silica-coated magnetic nanoparticles modified with γ-mercaptopropyltrimethoxysilane for fast and selective solid phase extraction of trace amounts of Cd, Cu, Hg, and Pb in environmental and biological samples prior to their determination by inductively coupled plasma mass spectrometry. Spectrochim. Acta Part B At. Spectrosc. 2008, 63, 437–444. [Google Scholar] [CrossRef]
- Dai, J.; de Cortalezzi, M.F. Influence of pH, ionic strength and natural organic matter concentration on a MIP-Fluorescent sensor for the quantification of DNT in water. Heliyon 2019, 5, e01922. [Google Scholar] [CrossRef] [Green Version]
- Cui, F.; Zhou, Z.; Zhou, H.S. Molecularly Imprinted Polymers and Surface Imprinted Polymers Based Electrochemical Biosensor for Infectious Diseases. Sensors 2020, 20, 996. [Google Scholar] [CrossRef] [Green Version]
- Duan, H.; Li, L.; Wang, X.; Wang, Y.; Li, J.; Luo, C. A sensitive and selective chemiluminescence sensor for the determination of dopamine based on silanized magnetic graphene oxide-molecularly imprinted polymer. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2015, 139, 374–379. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Ge, L.; Li, L.; Yan, M.; Ge, S.; Yu, J. Molecularly imprinted polymer grafted paper-based multi-disk micro-disk plate for chemiluminescence detection of pesticide. Biosens. Bioelectron. 2013, 50, 262–268. [Google Scholar] [CrossRef]
- Qiu, H.; Fan, L.; Li, X.; Li, L.; Sun, M.; Luo, C. A microflow chemiluminescence sensor for indirect determination of dibutyl phthalate by hydrolyzing based on biological recognition materials. J. Pharm. Biomed. Anal. 2013, 75, 123–129. [Google Scholar] [CrossRef]
- Cennamo, N.; Agostino, G.D.; Pesavento, M.; Zeni, L. High selectivity and sensitivity sensor based on MIP and SPR in tapered plastic optical fibers for the detection of l -nicotine. Sens. Actuators B Chem. 2014, 191, 529–536. [Google Scholar] [CrossRef]
- Gupta, B.D.; Shrivastav, A.M.; Usha, S.P. Surface Plasmon Resonance-Based Fiber Optic Sensors Utilizing Molecular Imprinting. Sensors 2016, 16, 1381. [Google Scholar] [CrossRef] [Green Version]
- Rahtuvanoğlu, A.; Akgönüllü, S.; Karacan, S.; Denizli, A. Biomimetic Nanoparticles Based Surface Plasmon Resonance Biosensors for Histamine Detection in Foods. Chemistryselect 2020, 5, 5683–5692. [Google Scholar] [CrossRef]
- Çakır, O.; Baysal, Z. Pesticide analysis with molecularly imprinted nanofilms using surface plasmon resonance sensor and LC-MS/MS: Comparative study for environmental water samples. Sensors Actuators B Chem. 2019, 297, 126764. [Google Scholar] [CrossRef]
- Özgür, E.; Topçu, A.A.; Yılmaz, E.; Denizli, A. Surface plasmon resonance based biomimetic sensor for urinary tract infections. Talanta 2020, 212, 120778. [Google Scholar] [CrossRef]
- Kamra, T.; Xu, C.; Montelius, L.; Schnadt, J.; Wijesundera, S.A.; Yan, M.; Ye, L. Photoconjugation of Molecularly Imprinted Polymer Nanoparticles for Surface-Enhanced Raman Detection of Propranolol. ACS Appl. Mater. Interfaces 2015, 7, 27479–27485. [Google Scholar] [CrossRef] [PubMed]
- Chang, L.; Ding, Y.; Li, X. Surface molecular imprinting onto silver microspheres for surface enhanc24 June 2013ed Raman scattering applications. Biosens. Bioelectron. 2013, 50, 106–110. [Google Scholar] [CrossRef] [PubMed]
- Xue, J.-Q.; Li, D.-W.; Qu, L.-L.; Long, Y.-T. Surface-imprinted core–shell Au nanoparticles for selective detection of bisphenol A based on surface-enhanced Raman scattering. Anal. Chim. Acta 2013, 777, 57–62. [Google Scholar] [CrossRef]
- Ren, X.; Li, X. Flower-like Ag coated with molecularly imprinted polymers as a surface-enhanced Raman scattering substrate for the sensitive and selective detection of glibenclamide. Anal. Methods 2020, 12, 2858–2864. [Google Scholar] [CrossRef] [PubMed]
- Liao, S.; Zhao, X.; Zhu, F.; Chen, M.; Wu, Z.; Song, X.; Yang, H.; Chen, X. Novel S, N-doped carbon quantum dot-based “off-on” fluorescent sensor for silver ion and cysteine. Talanta 2018, 180, 300–308. [Google Scholar] [CrossRef]
- Hsu, C.-Y.; Lee, M.-H.; Thomas, J.L.; Shih, C.-P.; Hung, T.-L.; Whang, T.-J.; Lin, H.-Y. Optical sensing of phenylalanine in urine via extraction with magnetic molecularly imprinted poly(ethylene-co-vinyl alcohol) nanoparticles. Nanotechnology 2015, 26, 305502. [Google Scholar] [CrossRef]
- Sergeyeva, T.A.; Chelyadina, D.S.; Gorbach, L.A.; Brovko, O.O.; Piletska, E.V.; Piletsky, S.A.; Sergeeva, L.M.; El’skaya, A.V. Colorimetric biomimetic sensor systems based on molecularly imprinted polymer membranes for highly-selective detection of phenol in environmental samples. Biopolym. Cell 2014, 30, 209–215. [Google Scholar] [CrossRef] [Green Version]
- Hu, Y.; Feng, S.; Gao, F.; Li-Chan, E.C.; Grant, E.; Lu, X. Detection of melamine in milk using molecularly imprinted polymers–surface enhanced Raman spectroscopy. Food Chem. 2015, 176, 123–129. [Google Scholar] [CrossRef]
- Guo, Z.; Chen, L.; Lv, H.; Yu, Z.; Zhao, B. Magnetic imprinted surface enhanced Raman scattering (MI-SERS) based ultrasensitive detection of ciprofloxacin from a mixed sample. Anal. Methods 2014, 6, 1627–1632. [Google Scholar] [CrossRef]
- Jiang, Y.; Wang, Y.; Meng, F.; Wang, B.; Cheng, Y.; Zhu, C. N-doped carbon dots synthesized by rapid microwave irradiation as highly fluorescent probes for Pb2+ detection. New J. Chem. 2015, 39, 3357–3360. [Google Scholar] [CrossRef]
- Cennamo, N.; De Maria, L.; D’Agostino, G.; Zeni, L.; Pesavento, M. Monitoring of Low Levels of Furfural in Power Transformer Oil with a Sensor System Based on a POF-MIP Platform. Sensors 2015, 15, 8499–8511. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sa-Nguanprang, S.; Phuruangrat, A.; Bunkoed, O. An optosensor based on a hybrid sensing probe of mesoporous carbon and quantum dots embedded in imprinted polymer for ultrasensitive detection of thiamphenicol in milk. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2022, 264, 120324. [Google Scholar] [CrossRef] [PubMed]
- Ahmadpour, H.; Hosseini, S.M.M. A solid-phase luminescence sensor based on molecularly imprinted polymer-CdSeS/ZnS quantum dots for selective extraction and detection of sulfasalazine in biological samples. Talanta 2019, 194, 534–541. [Google Scholar] [CrossRef]
- Feng, J.; Tao, Y.; Shen, X.; Jin, H.; Zhou, T.; Zhou, Y.; Hu, L.; Luo, D.; Mei, S.; Lee, Y.-I. Highly sensitive and selective fluorescent sensor for tetrabromobisphenol-A in electronic waste samples using molecularly imprinted polymer coated quantum dots. Microchem. J. 2018, 144, 93–101. [Google Scholar] [CrossRef]
- Wu, X.; Zhang, Z.; Li, J.; You, H.; Li, Y.; Chen, L. Molecularly imprinted polymers-coated gold nanoclusters for fluorescent detection of bisphenol A. Sensors Actuators B Chem. 2015, 211, 507–514. [Google Scholar] [CrossRef]
- Xu, J.; Zhang, R.; Liu, C.; Sun, A.; Chen, J.; Zhang, Z.; Shi, X. Highly Selective Electrochemiluminescence Sensor Based on Molecularly Imprinted-quantum Dots for the Sensitive Detection of Cyfluthrin. Sensors 2020, 20, 884. [Google Scholar] [CrossRef] [Green Version]
- Liu, Y.-Y.; Xu, X.; Xin, J.-W.; Ghulamb, M.; Fan, J.; Dong, X.; Qiu, L.-L.; Xue, M.; Meng, Z.-H. Molecularly imprinted colloidal array for the high-throughput screening of explosives. Chin. J. Anal. Chem. 2023, 51, 100215. [Google Scholar] [CrossRef]
- Altintas, Z.; Gittens, M.; Guerreiro, A.; Thompson, K.-A.; Walker, J.; Piletsky, S.; Tothill, I.E. Detection of Waterborne Viruses Using High Affinity Molecularly Imprinted Polymers. Anal. Chem. 2015, 87, 6801–6807. [Google Scholar] [CrossRef]
- Zaidi, S.A. Molecular imprinting: A useful approach for drug delivery. Mater. Sci. Energy Technol. 2020, 3, 72–77. [Google Scholar] [CrossRef]
- Wang, Q.; Zhang, D. A novel fluorescence sensing method based on quantum dot-graphene and a molecular imprinting technique for the detection of tyramine in rice wine. Anal. Methods 2018, 10, 3884–3889. [Google Scholar] [CrossRef]
- Batista, A.D.; Silva, W.R.; Mizaikoff, B. Molecularly imprinted materials for biomedical sensing. Med. Devices Sens. 2021, 4, e10166. [Google Scholar] [CrossRef]
- Yadav, A.K.; Verma, D.; Dalal, N.; Kumar, A.; Solanki, P.R. Molecularly imprinted polymer-based nanodiagnostics for clinically pertinent bacteria and virus detection for future pandemics. Biosens. Bioelectron. X 2022, 12, 100257. [Google Scholar] [CrossRef]
- Vidic, J.; Manzano, M.; Chang, C.-M.; Jaffrezic-Renault, N. Advanced biosensors for detection of pathogens related to livestock and poultry. Veter. Res. 2017, 48, 11. [Google Scholar] [CrossRef] [Green Version]
- Zhang, J.; Wang, Y.; Lu, X. Molecular imprinting technology for sensing foodborne pathogenic bacteria. Anal. Bioanal. Chem. 2021, 413, 4581–4598. [Google Scholar] [CrossRef]
- Tokonami, S.; Nakadoi, Y.; Takahashi, M.; Ikemizu, M.; Kadoma, T.; Saimatsu, K.; Dung, L.Q.; Shiigi, H.; Nagaoka, T. Label-Free and Selective Bacteria Detection Using a Film with Transferred Bacterial Configuration. Anal. Chem. 2013, 85, 4925–4929. [Google Scholar] [CrossRef]
- Castelli, F.; Conti, B.; Conte, U.; Puglisi, G. Effect of molecular weight and storage times on tolmetin release from poly-d,l-lactide microspheres to lipid model membrane. A calorimetric study. J. Control. Release 1996, 40, 277–284. [Google Scholar] [CrossRef]
- Hong, C.-C.; Chen, C.-P.; Horng, J.-C.; Chen, S.-Y. Point-of-care protein sensing platform based on immuno-like membrane with molecularly-aligned nanocavities. Biosens. Bioelectron. 2013, 50, 425–430. [Google Scholar] [CrossRef]
- Tawfik, S.M.; Elmasry, M.R.; Sharipov, M.; Azizov, S.; Lee, C.H.; Lee, Y.-I. Dual emission nonionic molecular imprinting conjugated polythiophenes-based paper devices and their nanofibers for point-of-care biomarkers detection. Biosens. Bioelectron. 2020, 160, 112211. [Google Scholar] [CrossRef]
- Orbay, S.; Kocaturk, O.; Sanyal, R.; Sanyal, A. Molecularly Imprinted Polymer-Coated Inorganic Nanoparticles: Fabrication and Biomedical Applications. Micromachines 2022, 13, 1464. [Google Scholar] [CrossRef]
- Díaz-Álvarez, M.; Martín-Esteban, A. Molecularly Imprinted Polymer-Quantum Dot Materials in Optical Sensors: An Overview of Their Synthesis and Applications. Biosensors 2021, 11, 79. [Google Scholar] [CrossRef] [PubMed]
- Cecchini, A.; Raffa, V.; Canfarotta, F.; Signore, G.; Piletsky, S.; MacDonald, M.P.; Cuschieri, A. In Vivo Recognition of Human Vascular Endothelial Growth Factor by Molecularly Imprinted Polymers. Nano Lett. 2017, 17, 2307–2312. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Peng, H.; Qin, Y.-T.; He, X.-W.; Li, W.-Y.; Zhang, Y.-K. Epitope Molecularly Imprinted Polymer Nanoparticles for Chemo-/Photodynamic Synergistic Cancer Therapy Guided by Targeted Fluorescence Imaging. ACS Appl. Mater. Interfaces 2020, 12, 13360–13370. [Google Scholar] [CrossRef] [PubMed]
- Dong, M.; Wang, Y.-W.; Peng, Y. Highly Selective Ratiometric Fluorescent Sensing for Hg2+ and Au3+, Respectively, in Aqueous Media. Org. Lett. 2010, 12, 5310–5313. [Google Scholar] [CrossRef]
- Feng, J.; Chen, X.; Han, Q.; Wang, H.; Lu, P.; Wang, Y. Naphthalene-based fluorophores: Synthesis characterization, and photophysical properties. J. Lumin. 2011, 131, 2775–2783. [Google Scholar] [CrossRef]
- Mehmandoust, M.; Erk, N.; Naser, M.; Soylak, M. Molecularly imprinted polymer film loaded on the metal–organic framework with improved performance using stabilized gold-doped graphite carbon nitride nanosheets for the single-step detection of Fenamiphos. Food Chem. 2023, 404, 134627. [Google Scholar] [CrossRef]
- Ghaani, M.; Büyüktaş, D.; Carullo, D.; Farris, S. Development of a New Electrochemical Sensor Based on Molecularly Imprinted Biopolymer for Determination of 4,4′-Methylene Diphenyl Diamine. Sensors 2023, 23, 46. [Google Scholar] [CrossRef]
- Zhou, T.; Feng, Y.; Zhou, L.; Tao, Y.; Luo, D.; Jing, T.; Shen, X.; Zhou, Y.; Mei, S. Selective and sensitive detection of tetrabromobisphenol-A in water samples by molecularly imprinted electrochemical sensor. Sensors Actuators B Chem. 2016, 236, 153–162. [Google Scholar] [CrossRef]
- George, A.; Cherian, A.R.; Benny, L.; Varghese, A.; Hegde, G. Surface-engineering of carbon fibre paper electrode through molecular imprinting technique towards electrochemical sensing of food additive in shrimps. Microchem. J. 2023, 184, 108155. [Google Scholar] [CrossRef]
- Lu, Z.; Li, S.; Li, Y.; Li, L.; Ma, H.; Wei, K.; Shi, C.; Sun, M.; Duan, R.; Wang, X.; et al. DFT-assisted design inspired by loofah-derived biomass carbon decorated CoFe-CoFe2O4 conjugated molecular imprinting strategy for hazardous thiamphenicol analysis in spiked food. Sensors Actuators B Chem. 2023, 374, 132852. [Google Scholar] [CrossRef]
- Ren, S.; Cheng, S.; Wang, Q.; Zheng, Z. Molecularly imprinted voltammetric sensor sensibilized by nitrogen-vacancy graphitized carbon nitride and Ag-MWCNTs towards the detection of acetaminophen. J. Mol. Recognit. 2022, 35, e2992. [Google Scholar] [CrossRef] [PubMed]
- Cetinkaya, A.; Kaya, S.I.; Atici, E.B.; Çorman, M.E.; Uzun, L.; Ozkan, S.A. A semi-covalent molecularly imprinted electrochemical sensor for rapid and selective detection of tiotropium bromide. Anal. Bioanal. Chem. 2022, 414, 8023–8033. [Google Scholar] [CrossRef]
- Sulym, I.; Cetinkaya, A.; Yence, M.; Çorman, M.E.; Uzun, L.; Ozkan, S.A. Novel electrochemical sensor based on molecularly imprinted polymer combined with L-His-MWCNTs@PDMS-5 nanocomposite for selective and sensitive assay of tetracycline. Electrochim. Acta 2022, 430, 141102. [Google Scholar] [CrossRef]
- Ghorbani, A.; Ganjali, M.R.; Ojani, R.; Raoof, J. Detection of Chloridazon in Aqueous Matrices Using a Nano- Sized Chloridazon-Imprinted Polymer-Based Voltammetric Sensor. Int. J. Electrochem. Sci. 2020, 15, 2913–2922. [Google Scholar] [CrossRef]
- Toro, M.J.U.; Marestoni, L.D.; Sotomayor, M.D.P.T. A new biomimetic sensor based on molecularly imprinted polymers for highly sensitive and selective determination of hexazinone herbicide. Sensors Actuators B Chem. 2015, 208, 299–306. [Google Scholar] [CrossRef]
- Bojdi, M.K.; Behbahani, M.; Najafi, M.; Bagheri, A.; Omidi, F.; Salimi, S. Selective and Sensitive Determination of Uranyl Ions in Complex Matrices by Ion Imprinted Polymers-Based Electrochemical Sensor. Electroanalysis 2015, 27, 2458–2467. [Google Scholar] [CrossRef]
- Alizadeh, T.; Mirzaee, S.; Rafiei, F. All-solid-state Cr(III)-selective potentiometric sensor based on Cr(III)-imprinted polymer nanomaterial/MWCNTs/carbon nanocomposite electrode. Int. J. Environ. Anal. Chem. 2017, 97, 1283–1297. [Google Scholar] [CrossRef]
- Alizadeh, T.; Ganjali, M.R.; Norouzi, P.; Zarejousheghani, M.; Zeraatkar, A. A novel high selective and sensitive para-nitrophenol voltammetric sensor, based on a molecularly imprinted polymer–carbon paste electrode. Talanta 2009, 79, 1197–1203. [Google Scholar] [CrossRef]
- Khadem, M.; Faridbod, F.; Norouzi, P.; Foroushani, A.R.; Ganjali, M.R.; Shahtaheri, S.J. Biomimetic electrochemical sensor based on molecularly imprinted polymer for dicloran pesticide determination in biological and environmental samples. J. Iran. Chem. Soc. 2016, 13, 2077–2084. [Google Scholar] [CrossRef]
- Alizadeh, T. Preparation of magnetic TNT-imprinted polymer nanoparticles and their accumulation onto magnetic carbon paste electrode for TNT determination. Biosens. Bioelectron. 2014, 61, 532–540. [Google Scholar] [CrossRef]
- Sarpong, K.A.; Xu, W.; Huang, W.; Yang, W. The Development of Molecularly Imprinted Polymers in the Clean-Up of Water Pollutants: A Review. Am. J. Anal. Chem. 2019, 10, 202–226. [Google Scholar] [CrossRef] [Green Version]
- Wong, A.; Foguel, M.V.; Khan, S.; de Oliveira, F.M.; Tarley, C.R.T.; Sotomayor, M.D. Development of an electrochemical sensor modified with mwcnt-cooh and mip for detection of diuron. Electrochimica Acta 2015, 182, 122–130. [Google Scholar] [CrossRef] [Green Version]
- Alizadeh, T.; Ganjali, M.R.; Akhoundian, M.; Norouzi, P. Voltammetric determination of ultratrace levels of cerium(III) using a carbon paste electrode modified with nano-sized cerium-imprinted polymer and multiwalled carbon nanotubes. Microchim. Acta 2016, 183, 1123–1130. [Google Scholar] [CrossRef]
- Khadem, M.; Faridbod, F.; Norouzi, P.; Foroushani, A.R.; Ganjali, M.R.; Yarahmadi, R.; Shahtaheri, S.J. Voltammetric Determination of Carbofuran Pesticide in Biological and Environmental Samples using a Molecularly Imprinted Polymer Sensor, a Multivariate Optimization. J. Anal. Chem. 2020, 75, 669–678. [Google Scholar] [CrossRef]
- Hande, P.; Samui, A.B.; Kulkarni, P.S. An Efficient Method for Determination of the Diphenylamine (Stabilizer) in Propellants by Molecularly Imprinted Polymer based Carbon Paste Electrochemical Sensor. Propellants Explos. Pyrotech. 2017, 42, 376–380. [Google Scholar] [CrossRef]
- Wu, S.; Li, K.; Dai, X.; Zhang, Z.; Ding, F.; Li, S. An ultrasensitive electrochemical platform based on imprinted chitosan/gold nanoparticles/graphene nanocomposite for sensing cadmium (II) ions. Microchem. J. 2020, 155, 104710. [Google Scholar] [CrossRef]
- Ghanei-Motlagh, M.; Taher, M.A. Magnetic silver(I) ion-imprinted polymeric nanoparticles on a carbon paste electrode for voltammetric determination of silver(I). Microchim. Acta 2017, 184, 1691–1699. [Google Scholar] [CrossRef]
- Soysal, M.; Muti, M.; Esen, C.; Gençdağ, K.; Aslan, A.; Erdem, A.; Karagözler, A.E. A Novel and Selective Methylene Blue Imprinted Polymer Modified Carbon Paste Electrode. Electroanalysis 2013, 25, 1278–1285. [Google Scholar] [CrossRef]
- Alizadeh, T. Comparison of different methodologies for integration of molecularly imprinted polymer and electrochemical transducer in order to develop a paraoxon voltammetric sensor. Thin Solid Films 2010, 518, 6099–6106. [Google Scholar] [CrossRef]
- Luo, X.; Huang, W.; Shi, Q.; Xu, W.; Luan, Y.; Yang, Y.; Wang, H.; Yang, W. Electrochemical sensor based on lead ion-imprinted polymer particles for ultra-trace determination of lead ions in different real samples. RSC Adv. 2017, 7, 16033–16040. [Google Scholar] [CrossRef] [Green Version]
- Rajabi, H.R.; Zarezadeh, A.; Karimipour, G. Porphyrin based nano-sized imprinted polymer as an efficient modifier for the design of a potentiometric copper carbon paste electrode. RSC Adv. 2017, 7, 14923–14931. [Google Scholar] [CrossRef] [Green Version]
- Ghanei-Motlagh, M.; Taher, M. Novel imprinted polymeric nanoparticles prepared by sol–gel technique for electrochemical detection of toxic cadmium(II) ions. Chem. Eng. J. 2017, 327, 135–141. [Google Scholar] [CrossRef]
- Goud, K.Y.; Satyanarayana, M.; Reddy, K.K.; Gobi, K.V. Development of highly selective electrochemical impedance sensor for detection of sub-micromolar concentrations of 5-Chloro-2,4-dinitrotoluene. J. Chem. Sci. 2016, 128, 763–770. [Google Scholar] [CrossRef]
- Khan, S.; Wong, A.; Zanoni, M.V.B.; Sotomayor, M.D.P.T. Electrochemical sensors based on biomimetic magnetic molecularly imprinted polymer for selective quantification of methyl green in environmental samples. Mater. Sci. Eng. C 2019, 103, 109825. [Google Scholar] [CrossRef]
- Singh, R.; Singh, M. Molecularly imprinted electrochemical sensor for highly selective and sensitive determination of artificial sweetener Acesulfame-K. Talanta Open 2023, 7, 100194. [Google Scholar] [CrossRef]
- Lu, Z.; Wei, K.; Ma, H.; Duan, R.; Sun, M.; Zou, P.; Yin, J.; Wang, X.; Wang, Y.; Wu, C.; et al. Bimetallic MOF synergy molecularly imprinted ratiometric electrochemical sensor based on MXene decorated with polythionine for ultra-sensitive sensing of catechol. Anal. Chim. Acta 2023, 1251, 340983. [Google Scholar] [CrossRef]
- Bonyadi, S.; Ghanbari, K. Application of molecularly imprinted polymer and ZnO nanoparticles as a novel electrochemical sensor for tartrazine determination. Microchem. J. 2023, 187, 108398. [Google Scholar] [CrossRef]
- Fatah, M.A.A.; El-Moghny, M.G.A.; El-Deab, M.S.; El Nashar, R.M. Application of molecularly imprinted electrochemical sensor for trace analysis of Metribuzin herbicide in food samples. Food Chem. 2023, 404, 134708. [Google Scholar] [CrossRef] [PubMed]
- Zhou, B.; Sheng, X.; Xie, H.; Zhou, S.; Huang, L.; Zhang, Z.; Zhu, Y.; Zhong, M. Molecularly Imprinted Electrochemistry Sensor Based on AuNPs/RGO Modification for Highly Sensitive and Selective Detection of Nitrofurazone. Food Anal. Methods 2023, 16, 709–720. [Google Scholar] [CrossRef]
- Salimonnafs, Y.; MemarMaher, B.; Amirkhani, L.; Derakhshanfard, F. Fabrication of a molecular imprinted composite and its application in the measurement of ceftriaxone in an electrochemical sensor. Int. J. Polym. Mater. Polym. Biomater. 2023, 72, 366–375. [Google Scholar] [CrossRef]
- Alizadeh, T.; Mousavi, Z. Molecularly imprinted polymer specific to creatinine complex with copper(II) ions for voltammetric determination of creatinine. Microchim. Acta 2022, 189, 393. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.; Xuan, X.; Li, M.; Li, C.; Li, P.; Li, H. Molecularly imprinted Ni-polyacrylamide-based electrochemical sensor for the simultaneous detection of dopamine and adenine. Anal. Chim. Acta 2022, 1202, 339689. [Google Scholar] [CrossRef] [PubMed]
- Karthika, P.; Shanmuganathan, S.; Viswanathan, S. Electrochemical sensor for picric acid by using molecularly imprinted polymer and reduced graphene oxide modified pencil graphite electrode. Proc. Indian Natl. Sci. Acad. 2022, 88, 263–276. [Google Scholar] [CrossRef]
- Elfadil, D.; Palmieri, S.; Silveri, F.; Della Pelle, F.; Sergi, M.; Del Carlo, M.; Amine, A.; Compagnone, D. Fast sonochemical molecularly imprinted polymer synthesis for selective electrochemical determination of maleic hydrazide. Microchem. J. 2022, 180, 107634. [Google Scholar] [CrossRef]
- Herrera-Chacón, A.; Cetó, X.; del Valle, M. Molecularly imprinted polymers—Towards electrochemical sensors and electronic tongues. Anal. Bioanal. Chem. 2021, 413, 6117–6140. [Google Scholar] [CrossRef]
- Svalova, T.S.; Saigushkina, A.A.; Verbitskiy, E.V.; Chistyakov, K.A.; Varaksin, M.V.; Rusinov, G.L.; Charushin, V.N.; Kozitsina, A.N. Rapid and sensitive determination of nitrobenzene in solutions and commercial honey samples using a screen-printed electrode modified by 1,3-/1,4-diazines. Food Chem. 2022, 372, 131279. [Google Scholar] [CrossRef]
- Cetó, X.; Saint, C.P.; Chow, C.W.; Voelcker, N.H.; Prieto-Simón, B. Electrochemical detection of N-nitrosodimethylamine using a molecular imprinted polymer. Sensors Actuators B Chem. 2016, 237, 613–620. [Google Scholar] [CrossRef]
- Miao, J.; Liu, A.; Wu, L.; Yu, M.; Wei, W.; Liu, S. Magnetic ferroferric oxide and polydopamine molecularly imprinted polymer nanocomposites based electrochemical impedance sensor for the selective separation and sensitive determination of dichlorodiphenyltrichloroethane (DDT). Anal. Chim. Acta 2020, 1095, 82–92. [Google Scholar] [CrossRef]
- Elshafey, R.; Radi, A.-E. Electrochemical impedance sensor for herbicide alachlor based on imprinted polymer receptor. J. Electroanal. Chem. 2018, 813, 171–177. [Google Scholar] [CrossRef]
- Buensuceso, C.E.; Tiu, B.D.B.; Lee, L.P.; Sabido, P.M.G.; Nuesca, G.M.; Caldona, E.B.; del Mundo, F.R.; Advincula, R.C. Electropolymerized-molecularly imprinted polymers (E-MIPS) as sensing elements for the detection of dengue infection. Anal. Bioanal. Chem. 2022, 414, 1347–1357. [Google Scholar] [CrossRef]
- Tang, W.; Yin, L.; Sempionatto, J.R.; Moon, J.; Teymourian, H.; Wang, J. Touch-Based Stressless Cortisol Sensing. Adv. Mater. 2021, 33, e2008465. [Google Scholar] [CrossRef] [PubMed]
- Mobed, A.; Hasanzadeh, M.; Hassanpour, S.; Saadati, A.; Agazadeh, M.; Mokhtarzadeh, A. An innovative nucleic acid based biosensor toward detection of Legionella pneumophila using DNA immobilization and hybridization: A novel genosensor. Microchem. J. 2019, 148, 708–716. [Google Scholar] [CrossRef]
- Mani, A.; Rajeev, M.; Anirudhan, T. Silver decorated silanized graphene oxide based molecularly surface imprinted electrochemical sensor for the trace level detection of L- Tryptophan. Mater. Chem. Phys. 2023, 299, 127445. [Google Scholar] [CrossRef]
- Charlier, H.; David, M.; Lahem, D.; Debliquy, M. Electrochemical Detection of Penicillin G Using Molecularly Imprinted Conductive Co-Polymer Sensor. Appl. Sci. 2022, 12, 7914. [Google Scholar] [CrossRef]
- Tertis, M.; Sîrbu, P.; Suciu, M.; Bogdan, D.; Pana, O.; Cristea, C.; Simon, I. An Innovative Sensor Based on Chitosan and Graphene Oxide for Selective and Highly-Sensitive Detection of Serotonin. Chemelectrochem 2022, 9, e202101328. [Google Scholar] [CrossRef]
- Diouf, A.; Bouchikhi, B.; El Bari, N. A nonenzymatic electrochemical glucose sensor based on molecularly imprinted polymer and its application in measuring saliva glucose. Mater. Sci. Eng. C 2019, 98, 1196–1209. [Google Scholar] [CrossRef]
- Oliveira, A.E.F.; Pereira, A.C.; Ferreira, L.F. Disposable electropolymerized molecularly imprinted electrochemical sensor for determination of breast cancer biomarker CA 15-3 in human serum samples. Talanta 2023, 252, 123819. [Google Scholar] [CrossRef]
- Pourhajghanbar, M.; Arvand, M.; Habibi, M.F. Surface imprinting by using bi-functional monomers on spherical template magnetite for selective detection of levodopa in biological fluids. Talanta 2023, 254, 124136. [Google Scholar] [CrossRef]
- Ben Hassine, A.; Raouafi, N.; Moreira, F.T. Novel biomimetic Prussian blue nanocubes-based biosensor for Tau-441 protein detection. J. Pharm. Biomed. Anal. 2023, 226, 115251. [Google Scholar] [CrossRef]
- Beiki, T.; Najafpour-Darzi, G.; Mohammadi, M.; Shakeri, M.; Boukherroub, R. Fabrication of a novel electrochemical biosensor based on a molecular imprinted polymer-aptamer hybrid receptor for lysozyme determination. Anal. Bioanal. Chem. 2023, 415, 899–911. [Google Scholar] [CrossRef]
- Ahmed, S.; Ansari, A.; Haidyrah, A.S.; Chaudhary, A.A.; Imran, M.; Khan, A. Hierarchical Molecularly Imprinted Inverse Opal-Based Platforms for Highly Selective and Sensitive Determination of Histamine. ACS Appl. Polym. Mater. 2022, 4, 2783–2793. [Google Scholar] [CrossRef]
- Alam, I.; Lertanantawong, B.; Sutthibutpong, T.; Punnakitikashem, P.; Asanithi, P. Molecularly Imprinted Polymer-Amyloid Fibril-Based Electrochemical Biosensor for Ultrasensitive Detection of Tryptophan. Biosensors 2022, 12, 291. [Google Scholar] [CrossRef] [PubMed]
- Dykstra, G.; Reynolds, B.; Smith, R.; Zhou, K.; Liu, Y. Electropolymerized Molecularly Imprinted Polymer Synthesis Guided by an Integrated Data-Driven Framework for Cortisol Detection. ACS Appl. Mater. Interfaces 2022, 14, 25972–25983. [Google Scholar] [CrossRef]
- Yeasmin, S.; Wu, B.; Liu, Y.; Ullah, A.; Cheng, L.-J. Nano gold-doped molecularly imprinted electrochemical sensor for rapid and ultrasensitive cortisol detection. Biosens. Bioelectron. 2022, 206, 114142. [Google Scholar] [CrossRef] [PubMed]
- Roushani, M.; Farokhi, S.; Rahmati, Z. Development of a dual-recognition strategy for the aflatoxin B1 detection based on a hybrid of aptamer-MIP using a Cu2O NCs/GCE. Microchem. J. 2022, 178, 107328. [Google Scholar] [CrossRef]
- Jalalvand, A.R. Fabrication of a novel molecularly imprinted biosensor assisted by multi-way calibration for simultaneous determination of cholesterol and cholestanol in serum samples. Chemom. Intell. Lab. Syst. 2022, 226, 104587. [Google Scholar] [CrossRef]
- Yang, J.C.; Cho, C.H.; Choi, D.Y.; Park, J.P.; Park, J. Microcontact surface imprinting of affinity peptide for electrochemical impedimetric detection of neutrophil gelatinase-associated lipocalin. Sensors Actuators B Chem. 2022, 364, 131916. [Google Scholar] [CrossRef]
- Rahmati, Z.; Roushani, M. SARS-CoV-2 virus label-free electrochemical nanohybrid MIP-aptasensor based on Ni3(BTC)2 MOF as a high-performance surface substrate. Microchim. Acta 2022, 189, 287. [Google Scholar] [CrossRef]
- Ferreira, N.S.; Carneiro, L.P.; Viezzer, C.; Almeida, M.J.; Marques, A.C.; Pinto, A.M.; Fortunato, E.; Sales, M.G.F. Passive direct methanol fuel cells acting as fully autonomous electrochemical biosensors: Application to sarcosine detection. J. Electroanal. Chem. 2022, 922, 116710. [Google Scholar] [CrossRef]
- Li, Y.; Luo, L.; Nie, M.; Davenport, A.; Li, Y.; Li, B.; Choy, K.-L. A graphene nanoplatelet-polydopamine molecularly imprinted biosensor for Ultratrace creatinine detection. Biosens. Bioelectron. 2022, 216, 114638. [Google Scholar] [CrossRef]
- Saxena, K.; Murti, B.T.; Yang, P.-K.; Malhotra, B.D.; Chauhan, N.; Jain, U. Fabrication of a Molecularly Imprinted Nano-Interface-Based Electrochemical Biosensor for the Detection of CagA Virulence Factors of H. pylori. Biosensors 2022, 12, 1066. [Google Scholar] [CrossRef]
- Cerqueira, S.M.; Fernandes, R.; Moreira, F.T.; Sales, M.G.F. Development of an electrochemical biosensor for Galectin-3 detection in point-of-care. Microchem. J. 2021, 164, 105992. [Google Scholar] [CrossRef]
- Roushani, M.; Zalpour, N. Impedimetric ultrasensitive detection of trypsin based on hybrid aptamer-2DMIP using a glassy carbon electrode modified by nickel oxide nanoparticle. Microchem. J. 2022, 172, 106955. [Google Scholar] [CrossRef]
- Balayan, S.; Chauhan, N.; Chandra, R.; Jain, U. Molecular imprinting based electrochemical biosensor for identification of serum amyloid A (SAA), a neonatal sepsis biomarker. Int. J. Biol. Macromol. 2022, 195, 589–597. [Google Scholar] [CrossRef] [PubMed]
- Pareek, S.; Jain, U.; Balayan, S.; Chauhan, N. Ultra-sensitive nano- molecular imprinting polymer-based electrochemical sensor for Follicle-Stimulating Hormone (FSH) detection. Biochem. Eng. J. 2022, 180, 108329. [Google Scholar] [CrossRef]
- Cardoso, A.R.; de Sá, M.; Sales, M.G.F. An impedimetric molecularly-imprinted biosensor for Interleukin-1β determination, prepared by in-situ electropolymerization on carbon screen-printed electrodes. Bioelectrochemistry 2019, 130, 107287. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gonçalves, M.D.L.; Truta, L.A.N.; Sales, M.G.F.; Moreira, F.T.C. Electrochemical Point-of Care (PoC) Determination of Interleukin-6 (IL-6) Using a Pyrrole (Py) Molecularly Imprinted Polymer (MIP) on a Carbon-Screen Printed Electrode (C-SPE). Anal. Lett. 2021, 54, 2611–2623. [Google Scholar] [CrossRef]
- Jain, U.; Soni, S.; Balhara, Y.P.S.; Khanuja, M.; Chauhan, N. Dual-Layered Nanomaterial-Based Molecular Pattering on Polymer Surface Biomimetic Impedimetric Sensing of a Bliss Molecule, Anandamide Neurotransmitter. ACS Omega 2020, 5, 10750–10758. [Google Scholar] [CrossRef]
- Ahmad, O.S.; Bedwell, T.S.; Esen, C.; Garcia-Cruz, A.; Piletsky, S.A. Molecularly Imprinted Polymers in Electrochemical and Optical Sensors. Trends Biotechnol. 2019, 37, 294–309. [Google Scholar] [CrossRef]
- Bossi, A.M.; Maniglio, D. BioMIPs: Molecularly imprinted silk fibroin nanoparticles to recognize the iron regulating hormone hepcidin. Microchim. Acta 2022, 189, 66. [Google Scholar] [CrossRef] [PubMed]
- Disley, J.; Gil-Ramírez, G.; Gonzalez-Rodriguez, J. Chitosan-Based Molecularly Imprinted Polymers for Effective Trapping of the Nerve Agent Simulant Dimethyl Methylphosphonate. ACS Appl. Polym. Mater. 2023, 5, 935–942. [Google Scholar] [CrossRef]












| S. No | Functional Monomer | Crosslinker | Initiator | Porogenic Solvents | Morphology | Polymerization Type | Reference |
|---|---|---|---|---|---|---|---|
| 1 | 4-Vinyl Pyridine, Methacrylic Acid (MAA), Itaconic Acid. N-Vinylimidazole, Allylthiourea, Acrylamide, N-Methacryloyl-(L)-Cysteine, 2-VinylPyridine | Ethylene glycol dimethacrylate (EGDMA), Divinyl Benzene | AIBN, 2-Hydroxyethyl Methacrylate, Lauryl Peroxide, and Benzoyl Peroxide | Acetone, Cyclohexanol | Monolith | Bulk polymerization | [26] |
| 2 | Dithizone, N-[3-(2-Aminoethylamino) Propyl] Trimethoxysilane, 3-Isocyanatopropyl Triethoxysilane | Tetraethoxysilane | Ammonia | - | Dendritic | Sol-gel process | [27] |
| 3 | N-Propylacryl Amide | N,N-Methylene-Bis-Acrylamide (Mbam) | Ammonium Sulfate | Microspheres | Surface grafting polymerization | [28] | |
| 4 | Chitosan | Epichlorohydrin | Microspheres | Suspension Polymerization | [29] | ||
| 5 | Acrylamide and β-Cyclodextrin | Epichlorohydrin | Ammonium persulfate | Emulsion polymerization | [30] | ||
| 6 | N-Methacryloyl-(L)-Cysteine | Metylenebis (Acrylamide) | Ammonium Persulfate | Membranes | Multi-step swelling polymerization | [31] | |
| 7 | 2-Methacryloylamido Histidine | Poly(Ethylene Glycol) Diacrylate | Ammonium Persulfate | Membranes | Multi-step swelling polymerization | [32] | |
| 8 | MAA, Divinyl benzene | EGDMA | Micro particles | Precipitation polymerization | [33] | ||
| 9 | O-amino phenol | Nanoparticles | Electro deposition | [34] | |||
| 10 | O-Phenylene diamine | Nanowires | Electro deposition | [35] |
| S. No. | Polymerization Type | Advantages | Limitation | References |
|---|---|---|---|---|
| 1 | Bulk | Cost-effective method. Ease in preparation. Better control over the size of MIP particles synthesized | Low selectivity and reproducibility. Use of time-consuming processes. Need an ample amount of eluent to remove the template. No control over the shape of MIPs generated. The MIP obtained requires grinding, which results in some irregularities in the shape of the particles. Requirement of the huge amount of porogens during the fabrication process. | [37,58] |
| 2 | Suspension | Spherical particles with high porosity are obtained by this method. | Due to the influence of the dispersing media, MIPs produced in this manner have poor recognition sites compared to other techniques. This method is suitable only for hydrophobic monomers and initiators. | [43,59,60] |
| 3 | Emulsion | Spherical MIPs are formed. The binding sites on the surface of the spherical MIPs are distributed evenly, and the reuse rate is high for MIPs. | Due to their strong polarity and hydrogen bond-forming capacity, the water molecules in the aqueous phase affect the interaction between the template and monomer, resulting in an impaired imprinting process. This polymerization technique’s precipitation and separation processes are complicated as they require demulsifiers and coagulants, which are challenging to purify in the end. These impurities affect the physical properties of the MIPs formed. | [38,40] |
| 4 | Precipitation | This process results in high-purity MIPs compared to synthetic approaches like emulsion and suspension polymerizations. Regular-shaped MIP beads are obtained in good yields. Easy and less time-consuming method. | The precipitation only occurs when the polymeric chains are large enough to be insoluble in the reaction mixture. There is a need for high-speed homogenization to form particles of uniform size. The particles formed in the reaction are affected by slight variations in several factors, including the polarity of the solvent, the reaction temperature, and the stirring rate. Thus, the reaction conditions are to be monitored efficiently. | [41,42] |
| 5 | Multi-step swelling | This method results in uniform and monodispersed spherical MIP particle. | This method requires sophisticated procedures that are time-consuming. More importantly, the swelling degree of the MIPs should be cautiously controlled. The swelling can negatively influence the recognition ability of the MIPs. Thus, the swelling property of MIPs needs to be thoroughly evaluated to avoid losing its memory effect. | [51,60] |
| 6 | Surface imprinting | The mass transfer rate and efficiency are increased because of the increasedexposure of recognition sites on the surface. This results in better adsorption and specific recognition capacity, making it more suitable for separation or sensing applications. The amount of eluent needed for removing the template is meager compared to other bulk techniques. | The surface imprinting process is complicated, with many process parameters involved in obtaining a uniform MIP film. Thus, this is a time-consuming and expensive process. | [46] |
| 7 | Electrochemical | Deposition of MIPs with a precise thickness on an electrode surface is possible. There is little or no requirement for eluents to remove the template molecules. Crosslinkers or initiators are not required. | This is an expensive polymerization technique. The optimization of the MIP coating process is a complicated and time-consuming process. For instance, a thin coating results in very few recognition and rebinding sites. On the other hand, the removal of templates becomes complex, resulting in poor rebinding of analytes in the case of thicker coatings. | [61] |
| Optical Sensor Material | The Physical Form of Sensors | Detection Method | Monomer | Target | Sample | LoD | Reference |
|---|---|---|---|---|---|---|---|
| MIP | Paper | UV-Visible | MAA + Polyethyleneimine | Cd (II) | Lake water | 1–100 ng/mL | [83] |
| MIP-C-dots | Film | Fluorescence | acrylic acid (AA) + methylacrylate (MA) | 2,4- dinitrotoluene | Lake and tap water | 1–15 ppm, 0.28 ppm | [84] |
| MIP-C-dots | Film | Fluorescence | APTES | Cetricine | Urine, Saliva | 0.5–500 ng/mL, 0.41 ng/mL | [85] |
| Silanizedmagneticgraphene-MIP | Capillary tube | Chemiluminescence | Acrylamide (AM) | Dopamine | Urine, dopaminehydrochloride injection | 8–200 ng/mL, 1.5 ng/mL | [86] |
| MIP/Chromatographypaper | Paper disk | Chemiluminescence | AM | 2,4-dichlorophenoxyaceticacid | Lake and tap water | 5 pM–10 μM, 1 pM | [87] |
| MIP-Magnetic NP | Nanoparticles | Chemiluminescence | MAA | Dibutyl phthalate | Juice | 3.84 × 10−8–2.08 × 10−5 M | [88] |
| MIP | Optical fiber | Surface Plasmon resonance | MAA | Furfural | Transformer oil | 9–30 ppb | [89] |
| MIP | Optical fiber | Surface Plasmon resonance | MAA | Profenofos | PBS | 2.5 × 10−6 μg/L | [90] |
| MIP | Nanoparticles | Surface Plasmon resonance | N-methacryloyl-(L)-histidinemethyl ester | Histamine | Cheese | 0.58 ng/L | [91] |
| MIP | Nanofilm | Surface Plasmon resonance | N-methacryloyl-(L)-tryptophan methylester | Carbofuran, dimethoate | River water | 7.11 (carbofuran); 8.37(dimethoate) ng/L | [92] |
| MIP-Ag NP | Film | Surface Plasmon resonance | N-methacryloyl-(L)-histidinemethyl ester | Escherichia coli | Urine | 15–1,500,000 CFU/mL | [93] |
| MIP | Nanoparticles | Raman scattering | MAA | Propranolol | Human Urine | 7.7 × 10−4 M | [94] |
| MIP-Au NP | Core-shell | Raman scattering | 3-(triethoxysilyl)propylisocyanate (TEPIC) | Bisphenol A | Surface water, plastic-bottled beverages | 2.2 × 10−6–10−4 M, 5.37 × 10−7 M | [95] |
| MIP-Ag | Core-shell | Raman scattering | AM | Glibenclamide | Water | 1 ng/mL–100 μg/mL | [96] |
| Au-MIP | Nanoparticles | Raman scattering | MAA, AM | 2,6-dichlorophenol | Water | 0.02 nM | [97] |
| MIP-Au NP | Fine particles | Raman scattering | MAA | Atrazine | Apple Juice | 0.0012 (SERS) mg/L | [98] |
| Magnetic MIP | Nanoparticles | Fluorescence, Raman scattering | Poly(ethylene-co-vinylalcohol) | Phenylalanine | Human urine | 7–100 (F); 5–800 μg/mL (RS) | [99] |
| MIP | Membrane | UV-Visible | Itaconic acid | Phenol | Drinking, natural, and wastewater | 50 nM–10 mM, 50 nM | [100] |
| MIP | Fine particles | Raman scattering | MAA | Melamine | Milk | 0.005–0.05 mM, 0.012 mM | [101] |
| MIP-Magnetic NP | Core-shell | Raman scattering | MAA | Ciprofloxacin | Fetal bovine serum | 10−7–10−4 M | [102] |
| MIP | Film | Surface Plasmon resonance | MAA | Histamine | Fish | 25 μg/L | [103] |
| MIP | Optical Fiber | Surface Plasmon resonance | MAA | L-nicotine | Ultrapure water | 1.86 × 10−4–10−3 M | [104] |
| MIP-QD | Nanocomposite | Fluorescence | APTES | Thiamphenicol | Urine | 0.04 μM | [105] |
| MIP-CdSeS/ZnSQD | Glass slide | Fluorescence | MAA | Sulfasalazine | Human plasma and urine | 0.02–1.5 μM, 0.0071 μM | [106] |
| MIP-QD | Composite | Fluorescence | APTES | Tetrabromobisphenol-A | Electronic waste | 1–60 ng/mL | [107] |
| MIP | Au Nanocomposite | Fluorescence | APTES | Bisphenol A | Seawater | 0.1–13 μM | [108] |
| MIP | Hollow Nanoparticles | Fluorescence | Acrylamide | λ-cyhalothrin | Canal water | 10.26–160 nM | [16] |
| CdTe QD-MIP | Composite | Fluorescence | Acrylamide | λ-cyhalothrin | River water | 0.1–16 μM | [109] |
| MIP | Colloidal array | MAA | Hexanitrohexaaziasowurtzitane; Hexahyro-1,3,5-triazine; 2,4,5-trinitro toluene; 2,4-dinitrotoluene; 2,6 dinitritolune; 1,3,5-trinitrobenzene | [110] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ramajayam, K.; Ganesan, S.; Ramesh, P.; Beena, M.; Kokulnathan, T.; Palaniappan, A. Molecularly Imprinted Polymer-Based Biomimetic Systems for Sensing Environmental Contaminants, Biomarkers, and Bioimaging Applications. Biomimetics 2023, 8, 245. https://doi.org/10.3390/biomimetics8020245
Ramajayam K, Ganesan S, Ramesh P, Beena M, Kokulnathan T, Palaniappan A. Molecularly Imprinted Polymer-Based Biomimetic Systems for Sensing Environmental Contaminants, Biomarkers, and Bioimaging Applications. Biomimetics. 2023; 8(2):245. https://doi.org/10.3390/biomimetics8020245
Chicago/Turabian StyleRamajayam, Kalaipriya, Selvaganapathy Ganesan, Purnimajayasree Ramesh, Maya Beena, Thangavelu Kokulnathan, and Arunkumar Palaniappan. 2023. "Molecularly Imprinted Polymer-Based Biomimetic Systems for Sensing Environmental Contaminants, Biomarkers, and Bioimaging Applications" Biomimetics 8, no. 2: 245. https://doi.org/10.3390/biomimetics8020245
APA StyleRamajayam, K., Ganesan, S., Ramesh, P., Beena, M., Kokulnathan, T., & Palaniappan, A. (2023). Molecularly Imprinted Polymer-Based Biomimetic Systems for Sensing Environmental Contaminants, Biomarkers, and Bioimaging Applications. Biomimetics, 8(2), 245. https://doi.org/10.3390/biomimetics8020245