Genetic Basis for Morphological Variation in the Zebrafish Danio rerio: Insights from a Low-Heterozygosity Line
Abstract
:1. Introduction
2. Materials and Methods
2.1. Fish Sources and Breeding
2.2. Experimental Design
2.3. Assessed Variables
2.4. Statistics
3. Results
3.1. Body and Embryo Mass
3.2. Total Length
3.3. Yolk/Chorion Ratio
3.4. Condition Factor
3.5. Specific Growth Rate
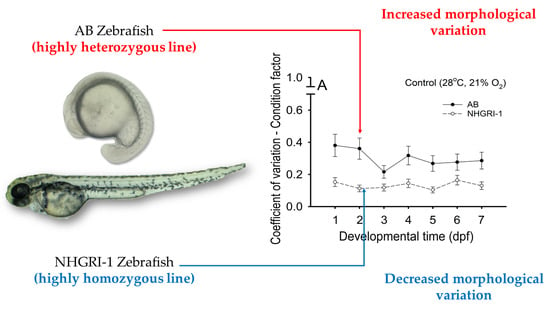
3.6. Coefficients of Variation
4. Discussion
4.1. Animal Models and Variation
4.2. The Zebrafish Model for Studying Mechanisms for Morphological Variation
4.3. Comparison of Variation in Wild-type and Inbred Zebrafish Embryos and Larvae
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- National Academies of Sciences; Engineering, and Medicine, Policy and Global Affairs; Committee on Science, Engineering, Medicine, and Public Policy; Board on Research Data and Information, Division on Engineering and Physical Sciences; Committee on Applied and Theoretical Statistics; Board on Mathematical Sciences and Analytics; Division on Earth and Life Studies, Nuclear and Radiation Studies Board; Division of Behavioral and Social Sciences and Education; Committee on National Statistics; Board on Behavioral, Cognitive, and Sensory Sciences; et al. Reproducibility and Replicability in Science; National Academies Press (US): Washington, DC, USA, 2019. [Google Scholar]
- Baker, M. 1500 scientists lift the lid on reproducibility. Nature 2016, 533, 452–454. [Google Scholar] [CrossRef] [PubMed]
- Ioannidis, J.P. Why most published research findings are false. PLoS Med. 2005, 2, e124. [Google Scholar] [CrossRef] [PubMed]
- Göpel, T.; Burggren, W.W. Insufficient reporting of experimental variables as a cause for nonreproducibility in animal physiology? A case study. Am. J. Physiol.-Regul. Integr. Comp. Physiol. 2022, 323, R363–R374. [Google Scholar] [CrossRef] [PubMed]
- Varga, Z.M.; Ekker, S.C.; Lawrence, C. Workshop Report: Zebrafish and Other Fish Models-Description of Extrinsic Environmental Factors for Rigorous Experiments and Reproducible Results. Zebrafish 2018, 15, 533–535. [Google Scholar] [CrossRef] [PubMed]
- Chan, J.C.; Houghton, A.B.; Bale, T.L. Strained in Planning Your Mouse Background? Using the HPA Stress Axis as a Biological Readout for Backcrossing Strategies. Neuropsychopharmacology 2017, 42, 1749–1751. [Google Scholar] [CrossRef] [PubMed]
- Crim, M.J.; Lawrence, C. A fish is not a mouse: Understanding differences in background genetics is critical for reproducibility. Lab Anim. 2021, 50, 19–25. [Google Scholar] [CrossRef] [PubMed]
- Franklin, C.L.; Ericsson, A.C. Microbiota and reproducibility of rodent models. Lab Anim. 2017, 46, 114–122. [Google Scholar] [CrossRef] [PubMed]
- Mandillo, S.; Tucci, V.; Hölter, S.M.; Meziane, H.; Banchaabouchi, M.A.; Kallnik, M.; Lad, H.V.; Nolan, P.M.; Ouagazzal, A.M.; Coghill, E.L.; et al. Reliability, robustness, and reproducibility in mouse behavioral phenotyping: A cross-laboratory study. Physiol. Genom. 2008, 34, 243–255. [Google Scholar] [CrossRef] [PubMed]
- Subramanian, S. Zebrafish as a model organism—Can a fish mimic human? J. Basic. Clin. Physiol. Pharmacol. 2023, 34, 559–575. [Google Scholar] [CrossRef] [PubMed]
- Chia, K.; Klingseisen, A.; Sieger, D.; Priller, J. Zebrafish as a model organism for neurodegenerative disease. Front. Mol. Neurosci. 2022, 15, 940484. [Google Scholar] [CrossRef]
- Ruchika; Sharma, A.; Saneja, A. Zebrafish as a powerful alternative model organism for preclinical investigation of nanomedicines. Drug Discov. Today 2022, 27, 1513–1522. [Google Scholar] [CrossRef] [PubMed]
- Chowdhury, S.; Saikia, S.K. Use of Zebrafish as a Model Organism to Study Oxidative Stress: A Review. Zebrafish 2022, 19, 165–176. [Google Scholar] [CrossRef] [PubMed]
- Suurväli, J.; Whiteley, A.R.; Zheng, Y.; Gharbi, K.; Leptin, M.; Wiehe, T. The Laboratory Domestication of Zebrafish: From Diverse Populations to Inbred Substrains. Mol. Biol. Evol. 2019, 37, 1056–1069. [Google Scholar] [CrossRef] [PubMed]
- Whiteley, A.R.; Bhat, A.; Martins, E.P.; Mayden, R.L.; Arunachalam, M.; Uusi-Heikkilä, S.; Ahmed, A.T.; Shrestha, J.; Clark, M.; Stemple, D.; et al. Population genomics of wild and laboratory zebrafish (Danio rerio). Mol. Ecol. 2011, 20, 4259–4276. [Google Scholar] [CrossRef] [PubMed]
- Audira, G.; Siregar, P.; Strungaru, S.A.; Huang, J.C.; Hsiao, C.D. Which Zebrafish Strains Are More Suitable to Perform Behavioral Studies? A Comprehensive Comparison by Phenomic Approach. Biology 2020, 9, 200. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Guo, N.; Lin, J.; Zhang, Y.; Chen, X.Q.; Li, S.; He, L.; Li, Q. Strain-dependent differential behavioral responses of zebrafish larvae to acute MK-801 treatment. Pharmacol. Biochem. Behav. 2014, 127, 82–89. [Google Scholar] [CrossRef] [PubMed]
- Mahabir, S.; Chatterjee, D.; Gerlai, R. Short exposure to low concentrations of alcohol during embryonic development has only subtle and strain- dependent effect on the levels of five amino acid neurotransmitters in zebrafish. Neurotoxicol. Teratol. 2018, 68, 91–96. [Google Scholar] [CrossRef]
- Meyer, B.M.; Froehlich, J.M.; Galt, N.J.; Biga, P.R. Inbred strains of zebrafish exhibit variation in growth performance and myostatin expression following fasting. Comp. Biochem. Physiol. A Mol. Integr. Physiol. 2013, 164, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Siregar, P.; Juniardi, S.; Audira, G.; Lai, Y.H.; Huang, J.C.; Chen, K.H.; Chen, J.R.; Hsiao, C.D. Method Standardization for Conducting Innate Color Preference Studies in Different Zebrafish Strains. Biomedicines 2020, 8, 271. [Google Scholar] [CrossRef] [PubMed]
- Volgin, A.D.; Yakovlev, O.A.; Demin, K.A.; de Abreu, M.S.; Alekseeva, P.A.; Friend, A.J.; Lakstygal, A.M.; Amstislavskaya, T.G.; Bao, W.; Song, C.; et al. Zebrafish models for personalized psychiatry: Insights from individual, strain and sex differences, and modeling gene x environment interactions. J. Neurosci. Res. 2019, 97, 402–413. [Google Scholar] [CrossRef] [PubMed]
- Wakamatsu, Y.; Ogino, K.; Hirata, H. Swimming capability of zebrafish is governed by water temperature, caudal fin length and genetic background. Sci. Rep. 2019, 9, 16307. [Google Scholar] [CrossRef] [PubMed]
- van den Bos, R.; Althuizen, J.; Tschigg, K.; Bomert, M.; Zethof, J.; Filk, G.; Gorissen, M. Early life exposure to cortisol in zebrafish (Danio rerio): Similarities and differences in behaviour and physiology between larvae of the AB and TL strains. Behav. Pharmacol. 2019, 30, 260–271. [Google Scholar] [CrossRef] [PubMed]
- Festing, M.F. Evidence should trump intuition by preferring inbred strains to outbred stocks in preclinical research. ILAR J. 2014, 55, 399–404. [Google Scholar] [CrossRef]
- Pathan, M.; Chaudhari, A.; Krishna, G.; Madhira, S. Inbred zebrafish lines: A genetic repository for zebrafish researchers. Indian J. Genet. Plant Breed. 2019, 79, 150–159. [Google Scholar] [CrossRef]
- Butler, M.G.; Iben, J.R.; Marsden, K.C.; Epstein, J.A.; Granato, M.; Weinstein, B.M. SNPfisher: Tools for probing genetic variation in laboratory-reared zebrafish. Development 2015, 142, 1542–1552. [Google Scholar] [CrossRef] [PubMed]
- Holden, L.A.; Wilson, C.; Heineman, Z.; Dobrinski, K.P.; Brown, K.H. An Interrogation of Shared and Unique Copy Number Variants Across Genetically Distinct Zebrafish Strains. Zebrafish 2019, 16, 29–36. [Google Scholar] [CrossRef] [PubMed]
- Lille-Langøy, R.; Karlsen, O.A.; Myklebust, L.M.; Goldstone, J.V.; Mork-Jansson, A.; Male, R.; Blumberg, B.; Stegeman, J.J.; Goksøyr, A. Sequence Variations in pxr (nr1i2) From Zebrafish (Danio rerio) Strains Affect Nuclear Receptor Function. Toxicol. Sci. 2019, 168, 28–39. [Google Scholar] [CrossRef]
- Chapman, F.A.; Fitz-Coy, S.A.; Thunberg, E.M.; Adams, C.M. United States of America Trade in Ornamental Fish. J. World Aquac. Soc. 1997, 28, 1–10. [Google Scholar] [CrossRef]
- LaFave, M.C.; Varshney, G.K.; Vemulapalli, M.; Mullikin, J.C.; Burgess, S.M. A defined zebrafish line for high-throughput genetics and genomics: NHGRI-1. Genetics 2014, 198, 167–170. [Google Scholar] [CrossRef] [PubMed]
- Meyers, J.R. Zebrafish: Development of a Vertebrate Model Organism. Curr. Protoc. Essent. Lab. Tech. 2018, 16, e19. [Google Scholar] [CrossRef]
- Tavares, B.; Santos Lopes, S. The importance of Zebrafish in biomedical research. Acta Med. Port. 2013, 26, 583–592. [Google Scholar] [CrossRef] [PubMed]
- Williams, S.Y.; Renquist, B.J. High Throughput Danio rerio Energy Expenditure Assay. J. Vis. Exp. 2016, 107, e53297. [Google Scholar] [CrossRef] [PubMed]
- Schneider, C.A.; Rasband, W.S.; Eliceiri, K.W. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 2012, 9, 671–675. [Google Scholar] [CrossRef] [PubMed]
- Ricker, W. Computation and Interpretation of Biological Statistics of Fish Populations. J. Fish. Res. Board Can. 1975, 191, 1–382. [Google Scholar]
- Brett, J.R.; Groves, T.D.D. 6—Physiological Energetics. In Fish Physiology; Hoar, W.S., Randall, D.J., Brett, J.R., Eds.; Academic Press: Cambridge, MA, USA, 1979; Volume 8, pp. 279–352. [Google Scholar]
- Holm, S. A Simple Sequentially Rejective Multiple Test Procedure. Scand. J. Stat. 1979, 6, 65–70. [Google Scholar]
- Aickin, M.; Gensler, H. Adjusting for multiple testing when reporting research results: The Bonferroni vs Holm methods. Am. J. Public Health 1996, 86, 726–728. [Google Scholar] [CrossRef] [PubMed]
- Irvin, H. A Report on the Statistical Properties of the Coefficient of Variation and Some Applications. Master’s Thesis, Utah State University, Logan, UT, USA, 1970. [Google Scholar]
- Gaertner, B.E.; Phillips, P.C. Caenorhabditis elegans as a platform for molecular quantitative genetics and the systems biology of natural variation. Genet. Res. 2010, 92, 331–348. [Google Scholar] [CrossRef]
- Barrière, A.; Félix, M.A. High local genetic diversity and low outcrossing rate in Caenorhabditis elegans natural populations. Curr. Biol. 2005, 15, 1176–1184. [Google Scholar] [CrossRef] [PubMed]
- Cutter, A.D. Nucleotide polymorphism and linkage disequilibrium in wild populations of the partial selfer Caenorhabditis elegans. Genetics 2006, 172, 171–184. [Google Scholar] [CrossRef] [PubMed]
- Sivasundar, A.; Hey, J. Population genetics of Caenorhabditis elegans: The paradox of low polymorphism in a widespread species. Genetics 2003, 163, 147–157. [Google Scholar] [CrossRef] [PubMed]
- Swan, K.A.; Curtis, D.E.; McKusick, K.B.; Voinov, A.V.; Mapa, F.A.; Cancilla, M.R. High-throughput gene mapping in Caenorhabditis elegans. Genome Res. 2002, 12, 1100–1105. [Google Scholar] [CrossRef] [PubMed]
- Wicks, S.R.; Yeh, R.T.; Gish, W.R.; Waterston, R.H.; Plasterk, R.H. Rapid gene mapping in Caenorhabditis elegans using a high density polymorphism map. Nat. Genet. 2001, 28, 160–164. [Google Scholar] [CrossRef] [PubMed]
- Stastna, J.J.; Snoek, L.B.; Kammenga, J.E.; Harvey, S.C. Genotype-dependent lifespan effects in peptone deprived Caenorhabditis elegans. Sci. Rep. 2015, 5, 16259. [Google Scholar] [CrossRef]
- van Wijk, M.H.; Davies, A.G.; Sterken, M.G.; Mathies, L.D.; Quamme, E.C.; Blackwell, G.G.; Riksen, J.A.G.; Kammenga, J.E.; Bettinger, J.C. Natural allelic variation modifies acute ethanol response phenotypes in wild strains of C. elegans. Alcohol. Clin. Exp. Res. 2023, 47, 1505–1517. [Google Scholar] [CrossRef] [PubMed]
- Kamkina, P.; Snoek, L.B.; Grossmann, J.; Volkers, R.J.; Sterken, M.G.; Daube, M.; Roschitzki, B.; Fortes, C.; Schlapbach, R.; Roth, A.; et al. Natural Genetic Variation Differentially Affects the Proteome and Transcriptome in Caenorhabditis elegans. Mol. Cell. Proteom. 2016, 15, 1670–1680. [Google Scholar] [CrossRef] [PubMed]
- Dolgin, E.S.; Félix, M.A.; Cutter, A.D. Hakuna Nematoda: Genetic and phenotypic diversity in African isolates of Caenorhabditis elegans and C. briggsae. Heredity 2008, 100, 304–315. [Google Scholar] [CrossRef]
- Kammenga, J.E.; Phillips, P.C.; De Bono, M.; Doroszuk, A. Beyond induced mutants: Using worms to study natural variation in genetic pathways. Trends Genet. 2008, 24, 178–185. [Google Scholar] [CrossRef] [PubMed]
- Johnson, T.E.; Hutchinson, E.W. Absence of strong heterosis for life span and other life history traits in Caenorhabditis elegans. Genetics 1993, 134, 465–474. [Google Scholar] [CrossRef] [PubMed]
- Graustein, A.; Gaspar, J.M.; Walters, J.R.; Palopoli, M.F. Levels of DNA polymorphism vary with mating system in the nematode genus caenorhabditis. Genetics 2002, 161, 99–107. [Google Scholar] [CrossRef] [PubMed]
- Jovelin, R.; Ajie, B.C.; Phillips, P.C. Molecular evolution and quantitative variation for chemosensory behaviour in the nematode genus Caenorhabditis. Mol. Ecol. 2003, 12, 1325–1337. [Google Scholar] [CrossRef] [PubMed]
- Berger, J.; Suzuki, T.; Senti, K.A.; Stubbs, J.; Schaffner, G.; Dickson, B.J. Genetic mapping with SNP markers in Drosophila. Nat. Genet. 2001, 29, 475–481. [Google Scholar] [CrossRef] [PubMed]
- Andersen, E.C.; Rockman, M.V. Natural genetic variation as a tool for discovery in Caenorhabditis nematodes. Genetics 2022, 220, iyab156. [Google Scholar] [CrossRef] [PubMed]
- Tinsley, M.C.; Blanford, S.; Jiggins, F.M. Genetic variation in Drosophila melanogaster pathogen susceptibility. Parasitology 2006, 132, 767–773. [Google Scholar] [CrossRef] [PubMed]
- Chakraborty, M.; VanKuren, N.W.; Zhao, R.; Zhang, X.; Kalsow, S.; Emerson, J.J. Hidden genetic variation shapes the structure of functional elements in Drosophila. Nat. Genet. 2018, 50, 20–25. [Google Scholar] [CrossRef] [PubMed]
- Mackay, T.F. Mutations and quantitative genetic variation: Lessons from Drosophila. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2010, 365, 1229–1239. [Google Scholar] [CrossRef] [PubMed]
- Shorter, J.; Couch, C.; Huang, W.; Carbone, M.A.; Peiffer, J.; Anholt, R.R.H.; Mackay, T.F.C. Genetic architecture of natural variation in Drosophila melanogaster aggressive behavior. Proc. Natl. Acad. Sci. USA 2015, 112, E3555–E3563. [Google Scholar] [CrossRef] [PubMed]
- Havula, E.; Ghazanfar, S.; Lamichane, N.; Francis, D.; Hasygar, K.; Liu, Y.; Alton, L.A.; Johnstone, J.; Needham, E.J.; Pulpitel, T.; et al. Genetic variation of macronutrient tolerance in Drosophila melanogaster. Nat. Commun. 2022, 13, 1637. [Google Scholar] [CrossRef] [PubMed]
- Palu, R.A.S.; Ong, E.; Stevens, K.; Chung, S.; Owings, K.G.; Goodman, A.G.; Chow, C.Y. Natural Genetic Variation Screen in Drosophila Identifies Wnt Signaling, Mitochondrial Metabolism, and Redox Homeostasis Genes as Modifiers of Apoptosis. G3 Genes Genomes Genet. 2019, 9, 3995–4005. [Google Scholar] [CrossRef] [PubMed]
- Frankham, R.; Briscoe, D.A.; Ballou, J.D. Introduction to Conservation Genetics; Cambridge University Press: Cambridge, UK, 2002. [Google Scholar]
- Jiang, P.P.; Lang, Q.L.; Fang, S.G.; Ding, P.; Chen, L.M. A genetic diversity comparison between captive individuals and wild individuals of Elliot’s Pheasant (Syrmaticus ellioti) using mitochondrial DNA. J. Zhejiang Univ. Sci. B 2005, 6, 413–417. [Google Scholar] [CrossRef]
- Mable, B.K.; Kilbride, E.; Viney, M.E.; Tinsley, R.C. Copy number variation and genetic diversity of MHC Class IIb alleles in an alien population of Xenopus laevis. Immunogenetics 2015, 67, 591–603. [Google Scholar] [CrossRef] [PubMed]
- Torres-Sánchez, M. Variation under domestication in animal models: The case of the Mexican axolotl. BMC Genom. 2020, 21, 827. [Google Scholar] [CrossRef] [PubMed]
- Yoshiki, A.; Ballard, G.; Perez, A.V. Genetic quality: A complex issue for experimental study reproducibility. Transgenic Res. 2022, 31, 413–430. [Google Scholar] [CrossRef] [PubMed]
- Poltorak, A.; Apalko, S.; Sherbak, S. Wild-derived mice: From genetic diversity to variation in immune responses. Mamm. Genome 2018, 29, 577–584. [Google Scholar] [CrossRef] [PubMed]
- Badanthadka, M.; D’Souza, L.; Salwa, F. Strain specific response of mice to IMQ-induced psoriasis. J. Basic Clin. Physiol. Pharmacol. 2021, 32, 959–968. [Google Scholar] [CrossRef] [PubMed]
- Caruso, A.; Marconi, M.A.; Scattoni, M.L.; Ricceri, L. Ultrasonic vocalizations in laboratory mice: Strain, age, and sex differences. Genes. Brain Behav. 2022, 21, e12815. [Google Scholar] [CrossRef] [PubMed]
- Corder, K.M.; Hoffman, J.M.; Sogorovic, A.; Austad, S.N. Behavioral comparison of the C57BL/6 inbred mouse strain and their CB6F1 siblings. Behav. Process. 2023, 207, 104836. [Google Scholar] [CrossRef] [PubMed]
- Khan, A.A.; Yurkovetskiy, L.; O’Grady, K.; Pickard, J.M.; de Pooter, R.; Antonopoulos, D.A.; Golovkina, T.; Chervonsky, A. Polymorphic Immune Mechanisms Regulate Commensal Repertoire. Cell Rep. 2019, 29, 541–550.e4. [Google Scholar] [CrossRef] [PubMed]
- Harmon, K.J.; Couper, L.L.; Lindner, V. Strain-dependent vascular remodeling phenotypes in inbred mice. Am. J. Pathol. 2000, 156, 1741–1748. [Google Scholar] [CrossRef] [PubMed]
- Locke, M.E.; Milojevic, M.; Eitutis, S.T.; Patel, N.; Wishart, A.E.; Daley, M.; Hill, K.A. Genomic copy number variation in Mus musculus. BMC Genom. 2015, 16, 497. [Google Scholar] [CrossRef] [PubMed]
- Loos, M.; Koopmans, B.; Aarts, E.; Maroteaux, G.; van der Sluis, S.; Verhage, M.; Smit, A.B. Within-strain variation in behavior differs consistently between common inbred strains of mice. Mamm. Genome 2015, 26, 348–354. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez-Algarra, F.; Seaborne, R.A.E.; Danson, A.F.; Yildizoglu, S.; Yoshikawa, H.; Law, P.P.; Ahmad, Z.; Maudsley, V.A.; Brew, A.; Holmes, N.; et al. Genetic variation at mouse and human ribosomal DNA influences associated epigenetic states. Genome Biol. 2022, 23, 54. [Google Scholar] [CrossRef] [PubMed]
- Angus, R.A.; Schultz, J. Clonal diversity in the unisexual fish poeciliopsis monacha-lucida: A tissue graft analysis. Evolution 1979, 33, 27–40. [Google Scholar] [CrossRef] [PubMed]
- Schultz, M.E.; Schultz, R.J. Differences in response to a chemical carcinogen within species and clones of the livebearing fish, Poeciliopsis. Carcinogenesis 1988, 9, 1029–1032. [Google Scholar] [CrossRef] [PubMed]
- Vrijenhoek, R.C.; Angus, R.A.; Schultz, R.J. Variation and heterozygosity in sexually vs. clonally reproducing populations of poeciliopsis. Evolution 1977, 31, 767–781. [Google Scholar] [CrossRef] [PubMed]
- Shen, Y.; Chalopin, D.; Garcia, T.; Boswell, M.; Boswell, W.; Shiryev, S.A.; Agarwala, R.; Volff, J.N.; Postlethwait, J.H.; Schartl, M.; et al. X. couchianus and X. hellerii genome models provide genomic variation insight among Xiphophorus species. BMC Genom. 2016, 17, 37. [Google Scholar] [CrossRef] [PubMed]
- Walter, R.; Hazlewood, L.; Kazianis, S. The Xiphophorus Genetic Stock Center Manual, 1st ed.; Kallman, K., Schartl, M., Eds.; Texas State University: San Marcos, TX, USA, 2006. [Google Scholar]
- Kirchmaier, S.; Naruse, K.; Wittbrodt, J.; Loosli, F. The genomic and genetic toolbox of the teleost medaka (Oryzias latipes). Genetics 2015, 199, 905–918. [Google Scholar] [CrossRef] [PubMed]
- Mrakovčič, M.; Haley, L.E. Inbreeding depression in the Zebra fish Brachydanio rerio (Hamilton Buchanan). J. Fish Biol. 1979, 15, 323–327. [Google Scholar] [CrossRef]
- Trevarrow, B.; Robison, B. Genetic backgrounds, standard lines, and husbandry of zebrafish. Methods Cell Biol. 2004, 77, 599–616. [Google Scholar] [CrossRef] [PubMed]
- Balik-Meisner, M.; Truong, L.; Scholl, E.H.; Tanguay, R.L.; Reif, D.M. Population genetic diversity in zebrafish lines. Mamm. Genome 2018, 29, 90–100. [Google Scholar] [CrossRef] [PubMed]
- Colón-Rodríguez, A.; Uribe-Salazar, J.M.; Weyenberg, K.B.; Sriram, A.; Quezada, A.; Kaya, G.; Jao, E.; Radke, B.; Lein, P.J.; Dennis, M.Y. Assessment of Autism Zebrafish Mutant Models Using a High-Throughput Larval Phenotyping Platform. Front. Cell Dev. Biol. 2020, 8, 586296. [Google Scholar] [CrossRef] [PubMed]
- Uribe-Salazar, J.M.; Kaya, G.; Sekar, A.; Weyenberg, K.; Ingamells, C.; Dennis, M.Y. Evaluation of CRISPR gene-editing tools in zebrafish. BMC Genom. 2022, 23, 12. [Google Scholar] [CrossRef]
- Venta, P.J.; Nguyen, A.K.; Senut, M.C.; Poulos, W.G.; Prukudom, S.; Cibelli, J.B. A 13-plex of tetra- and penta-STRs to identify zebrafish. Sci. Rep. 2020, 10, 3851. [Google Scholar] [CrossRef]
- Kendall, A.J.; Ahlstrom, E.; Moser, H. Early Life History Stages of Fishes and Their Characters; American Society of Ichthyologists and Herpetologists Special Publication: Glen Allen, VA, USA, 1984; pp. 11–22. [Google Scholar]
- Voesenek, C.J.; Muijres, F.T.; van Leeuwen, J.L. Biomechanics of swimming in developing larval fish. J. Exp. Biol. 2018, 221, jeb149583. [Google Scholar] [CrossRef] [PubMed]
- Lucas, A. Programming by early nutrition: An experimental approach. J. Nutr. 1998, 128, 401s–406s. [Google Scholar] [CrossRef] [PubMed]
- Rocha, F.; Dias, J.; Engrola, S.; Gavaia, P.; Geurden, I.; Dinis, M.T.; Panserat, S. Glucose overload in yolk has little effect on the long-term modulation of carbohydrate metabolic genes in zebrafish (Danio rerio). J. Exp. Biol. 2014, 217, 1139–1149. [Google Scholar] [CrossRef]
- Symonds, M.E.; Sebert, S.P.; Hyatt, M.A.; Budge, H. Nutritional programming of the metabolic syndrome. Nat. Rev. Endocrinol. 2009, 5, 604–610. [Google Scholar] [CrossRef] [PubMed]
- Waterland, R.A.; Jirtle, R.L. Early nutrition, epigenetic changes at transposons and imprinted genes, and enhanced susceptibility to adult chronic diseases. Nutrition 2004, 20, 63–68. [Google Scholar] [CrossRef] [PubMed]
- Diogo, P.; Martins, G.; Gavaia, P.; Pinto, W.; Dias, J.; Cancela, L.; Martínez-Páramo, S. Assessment of nutritional supplementation in phospholipids on the reproductive performance of zebrafish, Danio rerio (Hamilton, 1822). J. Appl. Ichthyol. 2015, 31, 3–9. [Google Scholar] [CrossRef]
- Jaya-Ram, A.; Kuah, M.-K.; Lim, P.-S.; Kolkovski, S.; Shu-Chien, A.C. Influence of dietary HUFA levels on reproductive performance, tissue fatty acid profile and desaturase and elongase mRNAs expression in female zebrafish Danio rerio. Aquaculture 2008, 277, 275–281. [Google Scholar] [CrossRef]
- Monteiro, J.F.; Martins, S.; Farias, M.; Costa, T.; Certal, A.C. The Impact of Two Different Cold-Extruded Feeds and Feeding Regimens on Zebrafish Survival, Growth and Reproductive Performance. J. Dev. Biol. 2018, 6, 15. [Google Scholar] [CrossRef] [PubMed]
- Tocher, D.R. Metabolism and Functions of Lipids and Fatty Acids in Teleost Fish. Rev. Fish. Sci. 2003, 11, 107–184. [Google Scholar] [CrossRef]
- Purushothaman, K.; Tan, J.K.H.; Lau, D.; Saju, J.M.; Thevasagayam, N.M.; Wee, C.L.; Vij, S. Feed Restriction Modulates Growth, Gut Morphology and Gene Expression in Zebrafish. Int. J. Mol. Sci. 2021, 22, 1814. [Google Scholar] [CrossRef] [PubMed]
- Dąbrowski, K. The feeding of fish larvae: Present «state of the art» and perspectives. Reprod. Nutr. Dévelop. 1984, 24, 807–833. [Google Scholar] [CrossRef]
- Gatesoupe, J.; Zambonino-Infante, J.; Cahu, C.; Bergot, P. Ontogeny, Development and Digestive Physiology of Fish Larvae. In Nutrition et Alimentation des Poissons et Crustaces; Guillaume, J., Kaushik, S., Bergot, P., Métailler, R., Eds.; Inra Editions: Versailles, France, 1999; pp. 249–264. [Google Scholar]
- Zambonino Infante, J.L.; Cahu, C.L. Ontogeny of the gastrointestinal tract of marine fish larvae. Comp. Biochem. Physiol. C Toxicol. Pharmacol. 2001, 130, 477–487. [Google Scholar] [CrossRef] [PubMed]
- Brown, M.E. Chapter IX—Experimental Studies on Growth. In The Physiology of Fishes, Brown, M.E., Ed.; Academic Press: Cambridge, MA, USA, 1957; pp. 361–400. [Google Scholar]
- Eaton, R.C.; Farley, R.D. Growth and the Reduction of Depensation of Zebrafish, Brachydanio rerio, Reared in the Laboratory. Copeia 1974, 1974, 204–209. [Google Scholar] [CrossRef]
- Ward, A.J.W.; Webster, M.M.; Hart, P.J.B. Intraspecific food competition in fishes. Fish Fish. 2006, 7, 231–261. [Google Scholar] [CrossRef]
- Parichy, D.M.; Elizondo, M.R.; Mills, M.G.; Gordon, T.N.; Engeszer, R.E. Normal table of postembryonic zebrafish development: Staging by externally visible anatomy of the living fish. Dev. Dyn. 2009, 238, 2975–3015. [Google Scholar] [CrossRef] [PubMed]
- Kimmel, C.B.; Ballard, W.W.; Kimmel, S.R.; Ullmann, B.; Schilling, T.F. Stages of embryonic development of the zebrafish. Dev. Dyn. 1995, 203, 253–310. [Google Scholar] [CrossRef] [PubMed]
- Burggren, W.W.; Bautista, G.M.; Coop, S.C.; Couturier, G.M.; Delgadillo, S.P.; García, R.M.; González, C.A.A. Developmental cardiorespiratory physiology of the air-breathing tropical gar, Atractosteus tropicus. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2016, 311, R689–R701. [Google Scholar] [CrossRef] [PubMed]
- Martínez, G.; Peña, E.; Martínez, R.; Camarillo, S.; Burggren, W.; Álvarez, A. Survival, Growth, and Development in the Early Stages of the Tropical Gar Atractosteus tropicus: Developmental Critical Windows and the Influence of Temperature, Salinity, and Oxygen Availability. Fishes 2021, 6, 5. [Google Scholar] [CrossRef]
- Pankhurst, N.W.; Munday, P.L. Effects of climate change on fish reproduction and early life history stages. Mar. Freshw. Res. 2011, 62, 1015–1026. [Google Scholar] [CrossRef]
- Kamei, H.; Lu, L.; Jiao, S.; Li, Y.; Gyrup, C.; Laursen, L.S.; Oxvig, C.; Zhou, J.; Duan, C. Duplication and diversification of the hypoxia-inducible IGFBP-1 gene in zebrafish. PLoS ONE 2008, 3, e3091. [Google Scholar] [CrossRef] [PubMed]
- Kamei, H.; Ding, Y.; Kajimura, S.; Wells, M.; Chiang, P.; Duan, C. Role of IGF signaling in catch-up growth and accelerated temporal development in zebrafish embryos in response to oxygen availability. Development 2011, 138, 777–786. [Google Scholar] [CrossRef] [PubMed]
- Kajimura, S.; Aida, K.; Duan, C. Insulin-like growth factor-binding protein-1 (IGFBP-1) mediates hypoxia-induced embryonic growth and developmental retardation. Proc. Natl. Acad. Sci. USA 2005, 102, 1240–1245. [Google Scholar] [CrossRef] [PubMed]
- Zhang, P.; Yao, Q.; Lu, L.; Li, Y.; Chen, P.J.; Duan, C. Hypoxia-inducible factor 3 is an oxygen-dependent transcription activator and regulates a distinct transcriptional response to hypoxia. Cell Rep. 2014, 6, 1110–1121. [Google Scholar] [CrossRef] [PubMed]
- Kopp, R.; Bauer, I.; Ramalingam, A.; Egg, M.; Schwerte, T. Prolonged hypoxia increases survival even in Zebrafish (Danio rerio) showing cardiac arrhythmia. PLoS ONE 2014, 9, e89099. [Google Scholar] [CrossRef] [PubMed]
- Bautista, G.M.; Padilla, P.; Burggren, W. Decreased physiological variation in the developing Zebrafish Danio rerio from a low heterozigosity line. In preparation.
- Biga, P.R.; Goetz, F.W. Zebrafish and giant danio as models for muscle growth: Determinate vs. indeterminate growth as determined by morphometric analysis. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2006, 291, R1327–R1337. [Google Scholar] [CrossRef] [PubMed]
- Biga, P.R.; Meyer, J. Growth hormone differentially regulates growth and growth-related gene expression in closely related fish species. Comp. Biochem. Physiol. A Mol. Integr. Physiol. 2009, 154, 465–473. [Google Scholar] [CrossRef] [PubMed]
- Parichy, D.M.; Turner, J.M. Zebrafish puma mutant decouples pigment pattern and somatic metamorphosis. Dev. Biol. 2003, 256, 242–257. [Google Scholar] [CrossRef] [PubMed]
- Georgakopoulou, E.; Sfakianakis, D.G.; Kouttouki, S.; Divanach, P.; Kentouri, M.; Koumoundouros, G. The influence of temperature during early life on phenotypic expression at later ontogenetic stages in sea bass. J. Fish Biol. 2007, 70, 278–291. [Google Scholar] [CrossRef]
- Klimogianni, A.; Koumoundouros, G.; Kaspiris, P.; Kentouri, M. Effect of temperature on the egg and yolk-sac larval development of common pandora, Pagellus erythrinus. Mar. Biol. 2004, 145, 1015–1022. [Google Scholar] [CrossRef]
- Martell, D.J.; Kieffer, J.D.; Trippel, E.A. Effects of temperature during early life history on embryonic and larval development and growth in haddock. J. Fish Biol. 2005, 66, 1558–1575. [Google Scholar] [CrossRef]
- de Ciechomski, J.D.; Sánchez, R.P.; Alespeiti, G.; Regidor, H. Estudio sobre el crecimiento en peso y factor de condición en larvas de anchoíta, Engraulis anchoita Hubbs & Marini. Variaciones regionales, estacionales y anuales. Rev. Investig. Desarro. Pesq. 1986, 5, 83–193. [Google Scholar]
- Westernhagen, H.V.; Rosenthal, H. On condition factor measurements in Pacific herring larvae. Helgoländer Meeresunters 1981, 34, 257–262. [Google Scholar] [CrossRef]




| TEMP (°C) | O2 (mg L−1)) | AGE (dpf) | Yolk/Chorion Ratio (%) | Body Mass (µg) | Embryo Mass (µg) | Total Length (mm) | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| WT AB | NHGRI-1 | WT AB | NHGRI-1 | WT AB | NHGRI-1 | WT AB | NHGRI-1 | |||
| 28 | 7.8 | 1 | 38 ± 1 | 43 ± 1 | 284 ± 8 | 276 ± 4 | 120 ± 3 | 110 ± 2 | 3.1 ± 0.1 | 2.5 ± 0.03 |
| 2 | 55.8 ± 2 | 51.6 ± 1.6 | 278 ± 10 | 266 ± 5 | 150 ± 4 | 120 ± 2 | 3.3 ± 0.1 | 2.8 ± 0.02 | ||
| 3 | 63.7 ± 2.2 | - | 305 ± 9 | 256 ± 4 | 180 ± 7 | 120 ± 2 | 3.9 ± 0.1 | 2.9 ± 0.03 | ||
| 4 | - | - | 315 ± 9 | 268 ± 3 | 230 ± 7 | 160 ± 2 | 4.2 ± 0.2 | 3.2 ± 0.04 | ||
| 5 | - | - | 349 ± 12 | 285 ± 4. | 280 ± 10 | 190 ± 3 | 4.2 ± 0.1 | 3.4 ± 0.03 | ||
| 6 | - | - | 329 ± 15 | 270 ± 6 | 290 ± 13 | 220 ± 5 | 4.2 ± 1 | 3.5 ± 0.04 | ||
| 7 | - | - | 314 ± 11 | 286 ± 4 | 300 ± 10 | 250 ± 4 | 4.4 ± 0.1 | 3.9 ± 0.05 | ||
| p < 0.001 | p < 0.001 | p < 0.001 | p < 0.001 | p < 0.001 | p < 0.001 | p < 0.001 | p < 0.001 | |||
| 23 | 7.8 | 1 | 47 ± 1.4 * | 39.9 ± 1.0 | 290 ± 7.2 | 215 ± 3 * | 120 ± 7 * | 90 ± 2. | 3.2 ± 0.10 | 2.3 ± .0.02 |
| 2 | 54.1 ± 1.3 | 54.9 ± 1.7 | 306 ± 7.3 | 215 ± 4 * | 130 ± 4 * | 130 ± 2 * | 3.4 ± 0.1 * | 2.4 ± 0.03 | ||
| 3 | 55.5 ± 2.2 | 53.8 ± 1.7 | 291 ± 8.5 | 227 ± 5 * | 130 ± 6 * | 130 ± 3 * | 3.7 ± 0.1 * | 2.9 ± 0.03 | ||
| 4 | 64.1 ± 2.3 | - | 297 ± 5.6 | 234 ± 5 * | 180 ± 6* | 160 ± 3 * | 3.8 ± 0.1 * | 3.2 ± 0.03 * | ||
| 5 | - | - | 306 ± 5.9 * | 253 ± 6 * | 210 ± 6 * | 200 ± 5 * | 3.8 ± 0.1 | 3.2 ± 0.04 | ||
| 6 | - | - | 339 ± 11.2 | 269 ± 4 | 280 ± 11 | 230 ± 4 * | 4.1 ± 0.1 | 3.2 ± 0.04 | ||
| 7 | - | - | 334 ± 10 | 279 ± 4. | 300 ± 10 * | 260 ± 4 * | 4.1 ± 0.1 | 3.4 ± 0.05 | ||
| p < 0.001 | p < 0.001 | p < 0.006 | p < 0.001 | p < 0.001 | p < 0.001 | p < 0.001 | p < 0.001 | |||
| 33 | 7.82 | 1 | 49.8 ± 2.2 * | 53 ± 2 * | 301 ± 7.9 | 243 ± 3.8 * | 120 ± 4 | 120 ± 2.1 * | 3.43 ± 0.1 | 2.7 ± 0.03 |
| 2 | - | - | 295 ± 7.2 | 300 ± 4.8 * | 170 ± 5 | 200 ± 3.3 | 3.98 ± 0.1 * | 3.1 ± 0.03 * | ||
| 3 | - | - | 317 ± 8.5 | 273 ± 5.8 * | 210 ± 6 * | 180 ± 3.7 * | 4.17 ± 0.1 * | 3.3 ± 0.03 * | ||
| 4 | - | - | 337 ± 11.3 * | 298 ± 3.8 * | 260 ± 9 | 220 ± 3.2 | 4.47 ± 0.1 * | 3.4 ± 0.04 * | ||
| 5 | - | - | 327 ± 9.3 | 300 ± 5.2 * | 280 ± 8 | 260 ± 4.5 | 4.51 ± 0.1 * | 3.5 ± 0.04 * | ||
| 6 | - | - | 337 ± 12.7 | 284 ± 3.4 | 300 ± 11 * | 250 ± 3.3 * | 4.59 ± 0.1 * | 3.7 ± 0.04 * | ||
| 7 | - | - | 293 ± 8.7 * | 276 ± 5.2 | 280 ± 8 | 270 ± 5.6 * | 4.65 ± 0.2 * | 4.2 ± 0.04 * | ||
| p < 0.001 | - | p < 0.026 | p < 0.001 | p < 0.001 | p < 0.001 | p < 0.001 | p < 0.001 | |||
| 28 | 4.9 | 1 | 38.1 ± 1.5 | 42.3 ± 2.2 | 294 ± 8.8 | 242 ± 5.7 * | 100 ± 7 * | 80 ± 2.2 * | 3.08 ± 0.1 | 2.1 ± 0.02 |
| 2 | 52 ± 1.6 | 55.1 ± 2.3 | 279 ± 7.7 | 233 ± 3.0 * | 110 ± 6 * | 100 ± 1.7 * | 3.30 ± 0.1 | 2.8 ± 0.03 | ||
| 3 | 43.9 ± 2.4 * | 50.5 ± 1.7 | 295 ± 7.8 | 229 ± 4.1 | 130 ± 8 * | 100 ± 1.9 * | 3.40 ± 0.2 * | 2.9 ± 0.04 * | ||
| 4 | - | - | 293 ± 7.6 | 209 ± 3.0 * | 170 ± 4 * | 120 ± 1.9 | 3.63 ± 0.2 * | 3 ± 0.04* | ||
| 5 | - | - | 297 ± 7.5 * | 245 ± 4.5 * | 190 ± 5 * | 150 ± 3.0 | 3.80 ± 0.1 * | 3.2 ± 0.04 * | ||
| 6 | - | - | 303 ± 7.0 | 260 ± 5.3 | 230 ± 9 * | 190 ± 4.0 * | 4.11 ± 0.1 | 3.4 ± 0.05 | ||
| 7 | - | - | 312 ± 9.7 | 275 ± 3.5 | 270 ± 9 * | 230 ± 2.8 * | 4.20 ± 0.1 | 3.6 ± 0.04 | ||
| p < 0.001 | p < 0.001 | p = 0.189 | p < 0.001 | p < 0.001 | p < 0.001 | p < 0.001 | p < 0.001 | |||
| 28 | 3.7 | 1 | 39.3 ± 1.5 | 46.2 ± 1.2 | 296 ± 7.0 | 219 ± 5 * | 110 ± 4 * | 80 ± 1.7 * | 3.0 ± 0.1 | 2.2 ± 0.02 |
| 2 | 54.1 ± 2 | 50.7 ± 1.8 | 288 ± 6.4 | 220 ± 4.1 * | 120 ± 6 * | 90 ± 2.0 * | 3.2 ± 0.1 | 2.5 ± 0.04 | ||
| 3 | 42.4 ± 2 * | 50.6 ± 3.4 | 291 ± 6.5 | 210 ± 3.4 | 140 ± 5 * | 90 ± 1.7 * | 3.3 ± 0.1 * | 2.9 ± 0.04 * | ||
| 4 | - | - | 302 ± 9.4 | 209 ± 3.6 * | 170 ± 1 * | 120 ± 2.4 | 3.5 ± 0.1 * | 3.0 ± 0.04 * | ||
| 5 | - | - | 293 ± 6.3 * | 250 ± 4.4 * | 180 ± 6 * | 150 ± 2.6 | 3.6 ± 0.1 * | 3.1 ± 0.05 * | ||
| 6 | - | - | 291 ± 7.2 | 263 ± 4.6 | 210 ± 7 * | 180 ± 3.2 * | 3.8 ± 0.1 | 3.3 ± 0.05 | ||
| 7 | - | - | 320 ± 9.6 | 266 ± 3.2 * | 260 ± 9 * | 210 ± 1.9 * | 4.0 ± 0.1 * | 3.3 ± 0.04 * | ||
| p < 0.001 | p = 0.264 | p = 0.217 | p < 0.001 | p < 0.001 | p < 0.001 | p < 0.001 | p < 0.001 | |||
| TEMP (°C) | O2 (mg L−1) | AGE (dpf) | Yolk/Chorion Ratio | Body Mass | Embryo Mass | Total Length | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| WT AB | NHGRI-1 | WT AB | NHGRI-1 | WT AB | NHGRI-1 | WT AB | NHGRI-1 | |||
| 28 | 7.8 | 1 | 0.08 ± 0.02 | 0.06 ± 0.01 * | 0.10 ± 0.02 | 0.05 ± 0.01 * | 0.08 ± .02 | 0.07 ± 0.01 | 0.09 ± 0.02 | 0.05 ± 0.01 * |
| 2 | 0.14 ± 0.02 | 0.12 ± 0.02 | 0.14 ± 0.03 | 0.07 ± 0.01 * | 0.11 ± 0.02 | 0.07 ± 0.01 * | 0.11 ± 0.02 | 0.03 ± 0.01 * | ||
| 3 | 0.13 ± 0.02 | - | 0.12 ± 0.02 | 0.06 ± 0.01 * | 0.14 ± 0.03 | 0.06 ± 0.01 * | 0.06 ± 0.01 | 0.04 ± 0.01 * | ||
| 4 | - | - | 0.11 ± 0.02 | 0.4 ± 0.02 * | 0.2 ± 0.03 | 0.05 ± 0.01 * | 0.08 ± 0.02 | 0.05 ± 0.01 * | ||
| 5 | - | - | 0.13 ± 0.02 | 0.05 ± 0.01 * | 0.14 ± 0.03 | 0.05 ± 0.01 * | 0.09 ± 0.02 | 0.04 ± 0.01 * | ||
| 6 | - | - | 0.18 ± 0.08 | 0.08 ± 0.02 * | 0.18 ± 0.03 | 0.09 ± 0.02 * | 0.09 ± 0.02 | 0.05 ± 0.01 * | ||
| 7 | - | - | 0.14 ± 0.02 | 0.05 ± 0.01 * | 0.14 ± 0.03 | 0.06 ± 0.01 * | 0.09 ± 0.02 | 0.05 ± 0.01 * | ||
| 23 | 7.8 | 1 | 0.11 ± 0.02 | 0.10 ± 0.02 | 0.10 ± 0.02 | 0.06 ± 0.01 * | 0.21 ± 0.04 | 0.08 ± 0.02 * | 0.13 ± 0.02 | 0.04 ± 0.01 * |
| 2 | 0.15 ± 0.03 | 0.12 ± 0.02 | 0.09 ± 0.02 | 0.07 ± 0.01 * | 0.13 ± 0.03 | 0.07 ± 0.01 * | 0.10 ± 0.02 | 0.04 ± .0.01 * | ||
| 3 | 0.15 ± 0.03 | 0.12 ± 0.02 | 0.11 ± 0.02 | 0.08 ± 0.01 * | 0.18 ± 0.03 | 0.09 ± 0.02 * | 0.08 ± 0.01 | 0.05 ± 0.01 * | ||
| 4 | 0.14 ± 0.03 | - | 0.07 ± 0.01 | 0.08 ± 0.01 | 0.13 ± 0.02 | 0.08 ± 0.01 * | 0.08 ± 0.02 | 0.04 ± 0.01 * | ||
| 5 | - | - | 0.07 ± 0.01 | 0.10 ± 0.02 | 0.10 ± 0.02 | 0.10 ± 0.02 | 0.13 ± 0.02 | 0.05 ± 0.01 * | ||
| 6 | - | - | 0.13 ± 0.02 | 0.06 ± 0.01 * | 0.15 ± 0.03 | 0.06 ± 0.01 * | 0.09 ± 0.02 | 0.05 ± 0.01 * | ||
| 7 | - | 0.12 ± 0.02 | 0.06 ± 0.01 * | 0.13 ± 0.02 | 0.06 ± 0.01 * | 0.10 ± 0.02 | 0.06 ± 0.01 * | |||
| 33 | 7.8 | 1 | 0.17 ± 0.03 | 0.20 ± 0.04 | 0.10 ± 0.02 | 0.06 ± 0.01 * | 0.14 ± 0.02 | 0.07 ± 0.01 * | 0.10 ± 0.02 | 0.04 ± 0.01 * |
| 2 | - | - | 0.09 ± 0.02 | 0.06 ± 0.01 * | 0.11 ± .0.02 | 0.06 ± 0.01 * | 0.05 ± 0.01 | 0.04 ± 0.01 | ||
| 3 | - | - | 0.10 ± 0.02 | 0.08 ± .0.02 | 0.11 ± 0.02 | 0.08 ± 0.01 * | 0.09 ± 0.02 | 0.04 ± 0.01 * | ||
| 4 | - | - | 0.13 ± 0.02 | 0.05 ± 0.01 * | 0.14 ± 0.03 | 0.06 ± 0.01 * | 0.09 ± 0.02 | 0.05 ± 0.01 * | ||
| 5 | - | - | 0.11 ± 0.02 | 0.07 ± 0.01 * | 0.10 ± 0.02 | 0.07 ± 0.01 * | 0.12 ± 0.02 | 0.04 ± 0.01 * | ||
| 6 | - | - | 0.15 ± 0.03 | 0.05 ± 0.01 * | 0.14 ± 0.03 | 0.05 ± 0.01 * | 0.10 ± 0.02 | 0.04 ± 0.01 * | ||
| 7 | 0.11 ± 0.02 | 0.07 ± 0.01 * | 0.11 ± 0.02 | 0.08 ± 0.01 * | 0.12 ± 0.02 | 0.04 ± 0.01 * | ||||
| 28 | 4.9 | 1 | 0.15 ± 0.03 | 0.20 ± 0.04 | 0.12 ± 0.02 | 0.09 ± 0.02 | 0.26 ± 0.05 | 0.10 ± 0.02 * | 0.07 ± 0.01 | 0.04 ± 0.01 * |
| 2 | 0.12 ± .02 | 0.16 ± 0.03 | 0.11 ± .0.02 | 0.05 ± 0.01 * | 0.19 ± 0.03 | 0.07 ± 0.01 * | 0.09 ± 0.02 | 0.04 ± 0.01 * | ||
| 3 | 0.21 ± 0.04 | 0.13 ± 0.02 * | 0.10 ± 0.02 | 0.07 ± 0.01 * | 0.24 ± 0.04 | 0.07 ± 0.01 * | 0.19 ± 0.04 | 0.10 ± 0.02 * | ||
| 4 | - | - | 0.10 ± 0.02 | 0.06 ± 0.01 * | 0.09 ± .0.02 | 0.06 ± 0.01 * | 0.18 ± 0.03 | 0.05 ± 0.01 * | ||
| 5 | - | - | 0.10 ± 0.02 | 0.07 ± 0.01 * | 0.10 ± 0.02 | 0.08 ± 0.01 | 0.11 ± 0.02 | 0.05 ± 0.01 * | ||
| 6 | - | - | 0.09 ± 0.02 | 0.08 ± 0.01 | 0.14 ± 0.03 | 0.08 ± 0.01 * | 0.07 ± 0.01 | 0.05 ± 0.01 | ||
| 7 | - | - | 0.12 ± 0.02 | 0.05 ± 0.01 * | 0.12 ± 0.02 | 0.05 ± 0.01 * | 0.09 ± 0.02 | 0.04 ± 0.01 * | ||
| 28 | 3.7 | 1 | 0.14 ± 0.03 | 0.10 ± 0.02 * | 0.09 ± 0.02 | 0.09 ± 0.02 | 0.14 ± 0.02 | 0.08 ± 0.02 * | 0.09 ± 0.02 | 0.04 ± 0.01 * |
| 2 | 0.14 ± 0.03 | 0.13 ± 0.02 | 0.09 ± 0.08 | 0.07 ± 0.01 | 0.19 ± 0.03 | 0.09 ± 0.02 * | 0.16 ± 0.03 | 0.07 ± 0.01 * | ||
| 3 | 0.18 ± 0.03 | 0.26 ± 0.05 | 0.09 ± 0.02 | 0.06 ± 0.01 * | 0.13 ± 0.02 | 0.07 ± 0.01 * | 0.15 ± 0.03 | 0.06 ± 0.01 * | ||
| 4 | - | - | 0.12 ± 0.02 | 0.07 ± 0.01 * | 0.22 ± 0.04 | 0.08 ± 0.01 * | 0.14 ± 0.03 | 0.05 ± 0.01 * | ||
| 5 | - | - | 0.08 ± 0.02 | 0.07 ± 0.01 | 0.12 ± 0.02 | 0.07 ± 0.01 * | 0.10 ± 0.02 | 0.06 ± 0.01 * | ||
| 6 | - | - | 0.10 ± 0.02 | 0.07 ± 0.01 * | 0.13 ± 0.02 | 0.07 ± 0.01 * | 0.12 ± 0.02 | 0.05 ± 0.01 * | ||
| 7 | - | - | 0.12 ± 0.02 | 0.04 ± 0.01 * | 0.13 ± 0.02 | 0.03 ± 0.01 * | 0.08 ± 0.01 | 0.04 ± 0.01 * | ||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Martinez-Bautista, G.; Padilla, P.; Burggren, W.W. Genetic Basis for Morphological Variation in the Zebrafish Danio rerio: Insights from a Low-Heterozygosity Line. Fishes 2024, 9, 164. https://doi.org/10.3390/fishes9050164
Martinez-Bautista G, Padilla P, Burggren WW. Genetic Basis for Morphological Variation in the Zebrafish Danio rerio: Insights from a Low-Heterozygosity Line. Fishes. 2024; 9(5):164. https://doi.org/10.3390/fishes9050164
Chicago/Turabian StyleMartinez-Bautista, Gil, Pamela Padilla, and Warren W. Burggren. 2024. "Genetic Basis for Morphological Variation in the Zebrafish Danio rerio: Insights from a Low-Heterozygosity Line" Fishes 9, no. 5: 164. https://doi.org/10.3390/fishes9050164
APA StyleMartinez-Bautista, G., Padilla, P., & Burggren, W. W. (2024). Genetic Basis for Morphological Variation in the Zebrafish Danio rerio: Insights from a Low-Heterozygosity Line. Fishes, 9(5), 164. https://doi.org/10.3390/fishes9050164