E2BMO: Facilitating User Interaction with a BioMimetic Ontology via Semantic Translation and Interface Design
Abstract
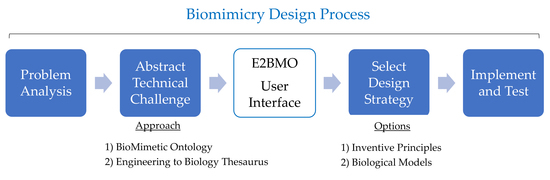
:1. Introduction
2. Materials and Methods
2.1. Selection of Tools
2.1.1. Engineering-to-Biology Thesaurus (E2B Thesaurus)
2.1.2. BioMimetic Ontology (BMO)
2.2. Merging of Tools
2.2.1. Simplified Knowledge Organization System (SKOS)
2.2.2. Creating a Combined User Interface
3. Results
3.1. E2BMO Website
3.1.1. BMO Trade-Off Concepts
3.1.2. E2B Functional Words
3.2. Case Study
3.2.1. BMO Trade-Offs
3.2.2. E2B Functional Words
4. Related Work
5. Discussion
- Expanding E2B thesaurus words through SKOS to include additional biological functions such as ‘pollinate, harvest, mate, etc.’.
- Translating the E2B Thesaurus Flow Terms to SKOS terms and tagging to inventive principles.
- Creating direct links to images or diagrams within the biological text data that may explain the design principle and help navigate the biological text in an interdisciplinary project.
- Analyzing the tool within an industrial context solving for real world technical challenges.
- Future work could potentially include incorporating data contributions from biologists into the E2BMO thus acting as an information sharing platform to a more diverse audience, possibly leading to potential collaboration and funding.
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| E2BMO | Engineering-to-BioMimetic Ontology |
| E2B | Engineering-to-Biology Thesaurus |
| BMO | BioMimetic Ontology |
| TRIZ | Teorija Reshenija Izobretateliskih Zadatch, which translates to Theory of Solving Problems Inventively |
| BFO | Basic Formal Ontology |
| IPs | Inventive Principles |
| SKOS | Simplified Knowledge Organization System |
| OWL | Web Ontology Language |
| RDF | Resource Description Framework |
| XML | Extensible Markup Language |
| SPARQL | a RDF query language |
| HTML | Hypertext markup language |
| JS | JavaScript |
| CSS | Cascading Style Sheets |
References
- Benyus, J.M. Spreading the Meme: A Biomimicry Primer. In Biomimicry Resource Handbook: A Seed Bank of Best Practice; Biomimicry 3.8: Missoula, MT, USA, 2013; pp. 7–19. ISBN 9781505634648. [Google Scholar]
- Biomimicry Global Design Challenge Toolbox: Case Studies. Available online: https://toolbox.biomimicry.org/references/case-studies/ (accessed on 12 October 2018).
- Mann, E.E.; Manna, D.; Mettetal, M.R.; May, R.M.; Dannemiller, E.M.; Chung, K.K.; Brennan, A.B.; Reddy, S.T. Surface micropattern limits bacterial contamination. Antimicrob. Resist. Infect. Control 2014, 3, 28. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Faure, E.; Lecomte, P.; Lenoir, S.; Vreuls, C.; Van De Weerdt, C.; Archambeau, C.; Martial, J.; Jérôme, C.; Duwez, A.-S.; Detrembleur, C. Sustainable and bio-inspired chemistry for robust antibacterial activity of stainless steel. J. Mater. Chem. 2011, 21, 7901–7904. [Google Scholar] [CrossRef]
- Zhang, X.; Zhang, Z.; Zhang, Y.; Wei, D.; Deng, Y. Route selection for emergency logistics management: A bio-inspired algorithm. Saf. Sci. 2013, 54, 87–91. [Google Scholar] [CrossRef]
- Kerbel, M.; Hoeller, N.; McKeag, T. REGEN Energy: The Power of Ants and Bees. Zygote Quarterly. 2012. Available online: https://zqjournal.org/editions/zq01.html (accessed on 12 October 2018).
- Fermanian Business & Economic Institute. Bioinspiration: An Economic Progress Report [Internet]; Point Loma Nazarene University: San Diego, CA, USA, 2013; Available online: http://www.magnefico.com/fileadmin/user_upload/Dokumente/PLNU_Bioinspiration_Da_Vinci_Index_A_Progress_Report_November_2013_Final.pdf (accessed on 28 March 2018).
- Wanieck, K.; Fayemi, P.-E.; Maranzana, N.; Zollfrank, C.; Jacobs, S. Biomimetics and its tools. Bioinspired Biomim. Nanobiomater. 2017, 6, 53–66. [Google Scholar] [CrossRef]
- Vattam, S.; Helms, M.; Goel, A.K. Biologically-Inspired Innovation in Engineering Design: A Cognitive Study; Georgia Institute of Technology: Atlanta, Georgia, 2007; p. 41. [Google Scholar]
- Helms, M.E.; Vattam, S.S.; Goel, A.K.; Yen, J.; Weissburg, M. Problem-Driven and Solution-Based Design. In Proceedings of the 28th Annual Conference of the Association for Computer Aided Design in Architecture (ACADIA), Minneapolis, MN, USA, 16–19 October 2008; pp. 94–101. [Google Scholar]
- Helms, M.; Vattam, S.S.; Goel, A.K. Biologically inspired design: Process and products. Des. Stud. 2009, 30, 606–622. [Google Scholar] [CrossRef]
- Fayemi, P.-E.; Wanieck, K.; Zollfrank, C.; Maranzana, N.; Aoussat, A. Biomimetics: Process, tools and practice. Bioinspir. Biomim. 2017, 12, 011002. [Google Scholar] [CrossRef] [PubMed]
- Massey, A.P.; Wallace, W.A. Understanding and facilitating group problem structuring and formulation: Mental representations, interaction, and representation aids. Decis. Support Syst. 1996, 17, 253–274. [Google Scholar] [CrossRef]
- Sartori, J.; Pal, U.; Chakrabarti, A. A methodology for supporting “transfer” in biomimetic design. Artif. Intell. Eng. Des. Anal. Manuf. 2010, 24, 483–506. [Google Scholar] [CrossRef]
- Baumeister, D. Biomimicry Resource Handbook: A Seed Bank of Best Practice; Biomimicry 3.8: Missoula, MT, USA, 2013; ISBN 9781505634648. [Google Scholar]
- Bogatyrev, N.R. Microfluidic Actuation in Living Organisms: A Biomimetic Catalogue. In Proceedings of the First European Conference on Microfluidics, Bologna, Italy, 10–12 December 2008; p. 175. [Google Scholar]
- Lenau, T. Biomimetics as a Design Methodology—Possibilities and Challenges. In Proceedings of the 17th International Conference on Engineering Design (ICED 09), Palo Alto, CA, USA, 24–27 August 2009. Volume 5 Design Methods and Tools (pt. 1). [Google Scholar]
- Cheong, H.; Chiu, I.; Shu, L.H.; Stone, R.B.; McAdams, D.A. Biologically Meaningful Keywords for Functional Terms of the Functional Basis. J. Mech. Des. 2011, 133, 021007. [Google Scholar] [CrossRef]
- Nagel, J.K.S.; Stone, R.B.; McAdams, D.A. An Engineering-to-Biology Thesaurus for Engineering Design. In Volume 5: 22nd International Conference on Design Theory and Methodology; Special Conference on Mechanical Vibration and Noise, Montreal, QC, Canada, 15–18 August 2010; ASME: New York, NY, USA, 2010; pp. 117–128. [Google Scholar]
- Kruiper, R.; Vincent, J.; Abraham, E.; Soar, R.; Konstas, I.; Chen-Burger, J.; Desmulliez, M. Towards a Design Process for Computer-Aided Biomimetics. Biomimetics 2018, 3, 14. [Google Scholar] [CrossRef]
- Autumn, K. How Gecko Toes Stick. Am. Sci. 2006, 94, 123–132. Available online: https://webdisk.lclark.edu/xythoswfs/webui/_xy-1594799_1-t_d9VVAITO (accessed on 12 October 2018). [CrossRef]
- Autumn, K.; Sitti, M.; Liang, Y.A.; Peattie, A.M.; Hansen, W.R.; Sponberg, S.; Kenny, T.W.; Fearing, R.; Israelachvili, J.N.; Full, R.J. Evidence for van der Waals adhesion in gecko setae. Proc. Natl. Acad. Sci. USA 2002, 99, 12252–12256. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hiller, U. Untersuchungen zum Feinbau und zur Funktion der Haftborsten von Reptilien. Z. Morphol. Tiere 1968, 62, 307–362. [Google Scholar] [CrossRef]
- Maderson, P.F.A. Keratinized Epidermal Derivatives as an Aid to Climbing in Gekkonid Lizards. Nature 1964, 203, 780–781. [Google Scholar] [CrossRef]
- Ruibal, R.; Ernst, V. The structure of the digital setae of lizards. J. Morphol. 1965, 117, 271–293. [Google Scholar] [CrossRef] [PubMed]
- Hansen, W.R.; Autumn, K. Evidence for self-cleaning in gecko setae. Proc. Natl. Acad. Sci. USA 2005, 102, 385–389. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Niewiarowski, P.H.; Stark, A.Y.; Dhinojwala, A. Sticking to the story: Outstanding challenges in gecko-inspired adhesives. J. Exp. Biol. 2016, 219, 912–919. [Google Scholar] [CrossRef] [PubMed]
- Stark, A.Y.; Badge, I.; Wucinich, N.A.; Sullivan, T.W.; Niewiarowski, P.H.; Dhinojwala, A. Surface wettability plays a significant role in gecko adhesion underwater. Proc. Natl. Acad. Sci. USA 2013, 110, 6340–6345. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kaiser, M.K.; Hashemi Farzaneh, H.; Lindemann, U. An approach to support searching for biomimetic solutions based on system characteristics and its environmental interactions. In DS 70: Proceedings of DESIGN 2012, the 12th International Design Conference, Dubrovnik, Croatia, 21–24 May 2012; pp. 969–978.
- Jacobs, S.R.; Nichol, E.C.; Helms, M.E. “Where Are We Now and Where Are We Going?” The BioM Innovation Database. J. Mech. Des. 2014, 136, 111101. [Google Scholar] [CrossRef]
- Vincent, J.F.V. The trade-off: A central concept for biomimetics. Bioinspired Biomim. Nanobiomater. 2017, 6, 67–76. [Google Scholar] [CrossRef]
- The OBO Foundry. Available online: http://www.obofoundry.org/ (accessed on 12 October 2018).
- Hirtz, J.; Stone, R.B.; McAdams, D.A.; Szykman, S.; Wood, K.L. A functional basis for engineering design: Reconciling and evolving previous efforts. Res. Eng. Des. 2002, 13, 65–82. [Google Scholar] [CrossRef] [Green Version]
- Altshuller, G.S. Creativity as an Exact Science: The Theory of the Solution of Inventive Problems; CRC Press LLC: Boca Raton, FL, USA, 1984; ISBN 9780677212302. [Google Scholar]
- Musen, M.A. The Protégé Project: A Look Back and a Look Forward. AI Matters 2015, 1, 4–12. [Google Scholar] [CrossRef] [PubMed]
- Miles, A.; Bechhofer, S.; SKOS Simple Knowledge Organization System Reference [Internet]. W3C Recommendation c2009. Available online: https://www.w3.org/TR/2009/REC-skos-reference-20090818/ (accessed on 28 March 2018).
- Baker, T.; Bechhofer, S.; Isaac, A.; Miles, A.; Schreiber, G.; Summers, E. Key choices in the design of Simple Knowledge Organization System (SKOS). Web Semant. Sci. Serv. Agents World Wide Web 2013, 20, 35–49. [Google Scholar] [CrossRef]
- Kozaki, K.; Mizoguchi, R. A Keyword Exploration for Retrieval from Biomimetics Databases. In Semantic Technology; Supnithi, T., Yamaguchi, T., Pan, J.Z., Wuwongse, V., Buranarach, M., Eds.; Springer International Publishing: Cham, Switzerland, 2015; Volume 8943, pp. 361–377. ISBN 978-3-319-15614-9. [Google Scholar]
- Yim, S.; Wilson, J.O. Development of an Ontology for Bio-Inspired Design using Description Logics. In Proceedings of the International Conference on Product Lifecycle Management, Seoul University, South Korea; July 2008. [Google Scholar]
- Gero, J.S.; Kannengiesser, U. A function-behavior-structure ontology of processes. AI Edam 2007, 21, 379–391. [Google Scholar] [CrossRef]
- Shu, L.H. A natural-language approach to biomimetic design. Artif. Intell. Eng. Des. Anal. Manuf. 2010, 24, 507–519. [Google Scholar] [CrossRef]
- Vincent, J.F.V.; Bogatyreva, O.A.; Bogatyrev, N.R.; Bowyer, A.; Pahl, A.-K. Biomimetics: Its practice and theory. J. R. Soc. Interface 2006, 3, 471–482. [Google Scholar] [CrossRef] [PubMed]









| Selected Inventive Principles | Selected Biological Function Correspondents |
|---|---|
| prepare for action | activate, catalyze, conserve, gauge, induce, learn, monitor, orient, observe, project |
| use asymmetry | concentrate, displace, deposit, elongate, fluctuate, filter, fold, hyperpolarize, segment, shorten |
| convert to dynamics | activate, articulate, bounce, catalyze, circulate, coil, conduct, contract, fold, fly, free, grow, jump, migrate, project, pump, twist, turn, roll, stretch, swim, swivel |
| prepare a defense | block, build, coil, concentrate, conserve, deposit, develop, envelope, excrete, expand, extend, induce, locate, obstruct, project, reduce, repel, repress, seal, synthesize, surround, tunnel, watch |
| hold the system in an equipotential state | catalyze, conserve, constrain, constrict, inhibit, maintain, oscillate, preserve, regulate, reduce, release, remain, respire, shorten, sustain |
| place or contain one object inside another | catch, clamp, clog, consume, cover, envelope, hold, inhale, intake, mate, roll, surround, suspend, tunnel, wrap |
| use feedback | acclimatize, activate, alter, communicate, constrain, constrict, evacuate, excrete, feed, fluctuate, gauge, halt, induce, learn, locate, maintain, migrate, monitor, orient, project, regulate, remain, repress, see, smell, sustain, trigger, watch |
| consolidate | bind, bond, breakdown, compress, concentrate, contract, decompose, filtrate, fold, fragment, fuse, orient, overlap, recombine, reduce, segment, shorten, wrap |
| use an intermediary | adhere, attach, bind, bond, catalyze, clamp, cling, clog, communicate, conduct, constrain, exchange, fluoresce, fuse, ionize, lactate, latch, lock, mate, obstruct, phosphorylate, photosynthesize, recombine, seal, see, smell, synthesize, transduce, watch |
| extract and separate | absorb, block, catch, concentrate, digest, deoxygenate, deaminate, decarboxylate, evacuate, excrete, filtrate, gate, intake, smell |
| change response threshold | activate, alter, catalyze, concentrate, constrain, extend, fluctuate, hold, inactivate, induce, react, reduce, regulate |
| change from static to dynamic field | alter, blend, build, circulate, displace, expand, free, grow, induce, liberate, react, release, scatter, |
| develop ability to predict | acclimatize, develop, gauge, learn, mark, monitor, observe, project, see, smell, watch |
| use periodic action | alternate, acclimatize, bounce, maintain, multiply, oscillate, replicate |
| change amplitude | alter, catalyze, concentrate, constrain, constrict, elongate, expand, extend, fluctuate, oscillate, reduce, regulate, shorten, stretch |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
McInerney, S.J.; Khakipoor, B.; Garner, A.M.; Houette, T.; Unsworth, C.K.; Rupp, A.; Weiner, N.; Vincent, J.F.V.; Nagel, J.K.S.; Niewiarowski, P.H. E2BMO: Facilitating User Interaction with a BioMimetic Ontology via Semantic Translation and Interface Design. Designs 2018, 2, 53. https://doi.org/10.3390/designs2040053
McInerney SJ, Khakipoor B, Garner AM, Houette T, Unsworth CK, Rupp A, Weiner N, Vincent JFV, Nagel JKS, Niewiarowski PH. E2BMO: Facilitating User Interaction with a BioMimetic Ontology via Semantic Translation and Interface Design. Designs. 2018; 2(4):53. https://doi.org/10.3390/designs2040053
Chicago/Turabian StyleMcInerney, Sarah J., Banafsheh Khakipoor, Austin M. Garner, Thibaut Houette, Colleen K. Unsworth, Ariana Rupp, Nicholas Weiner, Julian F. V. Vincent, Jacquelyn K. S. Nagel, and Peter H. Niewiarowski. 2018. "E2BMO: Facilitating User Interaction with a BioMimetic Ontology via Semantic Translation and Interface Design" Designs 2, no. 4: 53. https://doi.org/10.3390/designs2040053
APA StyleMcInerney, S. J., Khakipoor, B., Garner, A. M., Houette, T., Unsworth, C. K., Rupp, A., Weiner, N., Vincent, J. F. V., Nagel, J. K. S., & Niewiarowski, P. H. (2018). E2BMO: Facilitating User Interaction with a BioMimetic Ontology via Semantic Translation and Interface Design. Designs, 2(4), 53. https://doi.org/10.3390/designs2040053