Sub-Genotyping of Acanthamoeba T4 Complex: Experience from North India
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Isolates
2.2. Maintenance of Culture and Morphological Characterization
2.3. DNA Extraction
2.4. Molecular Characterization Using 16S
2.5. Phylogenetic Analysis
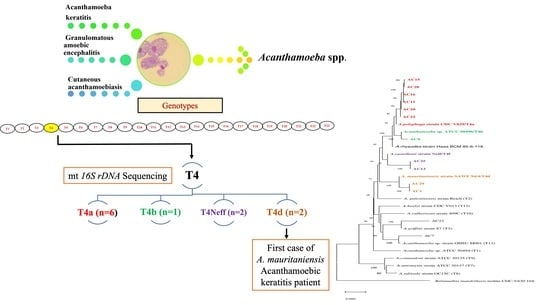
3. Results
3.1. Morphological Characterization of Study Isolates
3.2. Molecular Characterization and Phylogenetic Analysis
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Marciano-Cabral, F.; Cabral, G. Acanthamoeba spp. as Agents of Disease in Humans. Clin. Microbiol. Rev. 2003, 16, 273–307. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Khurana, S.; Sharma, M. Parasitic Keratitis—An under-Reported Entity. Trop. Parasitol. 2020, 10, 12–17. [Google Scholar] [CrossRef] [PubMed]
- Sharma, M.; Sudhan, S.S.; Sharma, S.; Megha, S.; Nada, R.; Khurana, S. Osteo-cutaneous acanthamoebiasis in a non-immunocompromised patient with a favorable outcome. Parasitol. Int. 2017, 66, 727–730. [Google Scholar] [CrossRef] [PubMed]
- Trabelsi, H.; Dendana, F.; Sellami, A.; Cheikhrouhou, F.; Neji, S.; Makni, F.; Ayadi, A. Pathogenic free-living amoebae: Epidemiology and clinical review. Pathol. Biol. 2012, 60, 399–405. [Google Scholar] [CrossRef]
- Schaumberg, D.A.; Snow, K.K.; Dana, M.R. The epidemic of Acanthamoeba keratitis: Where do we stand? Cornea 1998, 17, 3–10. [Google Scholar] [CrossRef]
- Sharma, S.; Garg, P.; Rao, G.N. Patient characteristics, diagnosis, and treatment of non-contact lens related Acanthamoeba keratitis. Br. J. Ophthalmol. 2000, 84, 1103–1108. [Google Scholar] [CrossRef] [Green Version]
- Saberi, R.; Nakhaei, M.; Fakhar, M.; Zarrinfar, H.; Sharifpour, A.; Hezarjaribi, H.Z. Molecular identification and genotyping of Acanthamoeba spp., in bronchoalveolar lavage fluid from immunocompetent patients with chronic respiratory disorders (CRD). Parasitol. Res. 2022, 121, 3013–3017. [Google Scholar] [CrossRef]
- Schuster, F.L.; Visvesvara, G.S. Free-living amoebae as opportunistic and non-opportunistic pathogens of humans and animals. Int. J. Parasitol. 2004, 34, 1001–1027. [Google Scholar] [CrossRef]
- Ledee, D.R.; Iovieno, A.; Miller, D.; Mandal, N.; Diaz, M.; Fell, J.; Fini, M.E.; Alfonso, E.C. Molecular Identification of T4 and T5 Genotypes in Isolates from Acanthamoeba Keratitis Patients. J. Clin. Microbiol. 2009, 47, 1458–1462. [Google Scholar] [CrossRef] [Green Version]
- Fuerst, P.A. Insights from the DNA databases: Approaches to the phylogenetic structure of Acanthamoeba. Exp. Parasitol. 2014, 145, S39–S45. [Google Scholar] [CrossRef]
- Khan, N.A.; Jarroll, E.L.; Paget, T.A. Molecular and Physiological Differentiation Between Pathogenic and Nonpathogenic Acanthamoeba. Curr. Microbiol. 2002, 45, 197–202. [Google Scholar] [CrossRef]
- Malavin, S.; Shmakova, L. Isolates from ancient permafrost help to elucidate species boundaries in Acanthamoeba castellanii complex (Amoebozoa: Discosea). Eur. J. Protistol. 2020, 73, 125671. [Google Scholar] [CrossRef] [PubMed]
- Diehl, M.L.N.; Paes, J.; Rott, M.B. Genotype distribution of Acanthamoeba in keratitis: A systematic review. Parasitol. Res. 2021, 120, 3051–3063. [Google Scholar] [CrossRef] [PubMed]
- Megha, K.; Sharma, M.; Sharma, C.; Gupta, A.; Sehgal, R.; Khurana, S. Evaluation of in vitro activity of five antimicrobial agents on Acanthamoeba isolates and their toxicity on human corneal epithelium. Eye 2021, 36, 1911–1917. [Google Scholar] [CrossRef]
- Pussard, M.; Pons, R. Morphologie de La Paroi Kystique et Taxonomie Du Genre Acanthamoeba (Protozoa, Amoebida). Protistologica 1977, 13, 557–598. [Google Scholar]
- Booton, G.C.; Visvesvara, G.S.; Byers, T.J.; Kelly, D.J.; Fuerst, P.A. Identification and Distribution of Acanthamoeba Species Genotypes Associated with Nonkeratitis Infections. J. Clin. Microbiol. 2005, 43, 1689–1693. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fuerst, P.A.; Booton, G.C.; Crary, M. Phylogenetic Analysis and the Evolution of the 18S rRNA Gene Typing System of Acanthamoeba. J. Eukaryot. Microbiol. 2015, 62, 69–84. [Google Scholar] [CrossRef]
- Megha, K.; Sehgal, R.; Khurana, S. Genotyping of Acanthamoeba spp. isolated from patients with granulomatous amoebic encephalitis. Indian J. Med. Res. 2018, 148, 456–459. [Google Scholar] [CrossRef]
- Megha, K.; Sharma, M.; Gupta, A.; Sehgal, R.; Khurana, S. Microbiological diagnosis of Acanthamoebic keratitis: Experience from tertiary care center of North India. Diagn. Microbiol. Infect. Dis. 2021, 100, 115339. [Google Scholar] [CrossRef] [PubMed]
- Megha, K.; Gupta, A.; Sehgal, R.; Khurana, S. An Improvised Medium for Axenic Cultivation of Acanthamoeba spp. Indian J. Med. Microbiol. 2017, 35, 597–599. [Google Scholar] [CrossRef]
- Ledee, D.R.; Booton, G.C.; Awwad, M.H.; Sharma, S.; Aggarwal, R.K.; Niszl, I.A.; Markus, M.B.; Fuerst, P.A.; Byers, T.J. Advantages of Using Mitochondrial 16S rDNA Sequences to Classify Clinical Isolates of Acanthamoeba. Investig. Opthalmology Vis. Sci. 2003, 44, 1142–1149. [Google Scholar] [CrossRef] [Green Version]
- Johnson, M.; Zaretskaya, I.; Raytselis, Y.; Merezhuk, Y.; McGinnis, S.; Madden, T.L. NCBI BLAST: A better web interface. Nucleic Acids Res. 2008, 36, W5–W9. [Google Scholar] [CrossRef] [PubMed]
- Larkin, M.A.; Blackshields, G.; Brown, N.P.; Chenna, R.; McGettigan, P.A.; McWilliam, H.; Valentin, F.; Wallace, I.M.; Wilm, A.; Lopez, R.; et al. Clustal W and Clustal X version 2.0. Bioinformatics 2007, 23, 2947–2948. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA6: Molecular Evolutionary Genetics Analysis Version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fuerst, P.; Booton, G. Species, Sequence Types and Alleles: Dissecting Genetic Variation in Acanthamoeba. Pathogens 2020, 9, 534. [Google Scholar] [CrossRef] [PubMed]
- Rayamajhee, B.; Sharma, S.; Willcox, M.; Henriquez, F.L.; Rajagopal, R.N.; Shrestha, G.S.; Subedi, D.; Bagga, B.; Carnt, N. Assessment of genotypes, endosymbionts and clinical characteristics of Acanthamoeba recovered from ocular infection. BMC Infect. Dis. 2022, 22, 757. [Google Scholar] [CrossRef] [PubMed]
- Prithiviraj, S.R.; Rajapandian, S.G.K.; Gnanam, H.; Gunasekaran, R.; Mariappan, P.; Singh, S.S.; Prajna, L. Clinical presentations, genotypic diversity and phylogenetic analysis of Acanthamoeba species causing keratitis. J. Med. Microbiol. 2020, 69, 87–95. [Google Scholar] [CrossRef]
- Corsaro, D. Update on Acanthamoeba phylogeny. Parasitol. Res. 2020, 119, 3327–3338. [Google Scholar] [CrossRef]
- Martín-Pérez, T.; Criado-Fornelio, A.; Martínez, J.; Blanco, M.; Fuentes, I.; Pérez-Serrano, J. Isolation and molecular characterization of Acanthamoeba from patients with keratitis in Spain. Eur. J. Protistol. 2017, 61, 244–252. [Google Scholar] [CrossRef]
- Rahman, M.; Yagita, K.; Kobayashi, A.; Oikawa, Y.; Hussein, A.; Matsumura, T.; Tokoro, M. Genetic Characterization of Clinical Acanthamoeba Isolates from Japan using Nuclear and Mitochondrial Small Subunit Ribosomal RNA. Korean J. Parasitol. 2013, 51, 401–411. [Google Scholar] [CrossRef]
- Putaporntip, C.; Kuamsab, N.; Nuprasert, W.; Rojrung, R.; Pattanawong, U.; Tia, T.; Yanmanee, S.; Jongwutiwes, S. Analysis of Acanthamoeba genotypes from public freshwater sources in Thailand reveals a new genotype, T23 Acanthamoeba bangkokensis sp. nov. Sci. Rep. 2021, 11, 17290. [Google Scholar] [CrossRef] [PubMed]
- Spotin, A.; Moslemzadeh, H.R.; Mahami-Oskouei, M.; Ahmadpour, E.; Niyyati, M.; Hejazi, S.H.; Memari, F.; Noori, J. Phylogeography, genetic variability and structure of Acanthamoeba metapopulations in Iran inferred by 18S ribosomal RNA sequences: A systematic review and meta-analysis. Asian Pac. J. Trop. Med. 2017, 10, 855–863. [Google Scholar] [CrossRef] [PubMed]
- Chelkha, N.; Jardot, P.; Moussaoui, I.; Levasseur, A.; La Scola, B.; Colson, P. Core gene-based molecular detection and identification of Acanthamoeba species. Sci. Rep. 2020, 10, 1583. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stothard, D.R.; Schroeder-Diedrich, J.M.; Awwad, M.H.; Gast, R.J.; Ledee, D.; Rodriguez-Zaragoza, S.; Dean, C.L.; Fuerst, P.; Byers, T.J. The Evolutionary History of the Genus Acanthamoeba and the Identification of Eight New 18S rRNA Gene Sequence Types. J. Eukaryot. Microbiol. 1998, 45, 45–54. [Google Scholar] [CrossRef] [PubMed]
- Xuan, Y.-H.; Chung, B.-S.; Hong, Y.-C.; Kong, H.-H.; Hahn, T.-W.; Chung, D.-I. Keratitis by Acanthamoeba triangularis: Report of Cases and Characterization of Isolates. Korean J. Parasitol. 2008, 46, 157–164. [Google Scholar] [CrossRef] [PubMed]
- Coronado-Velázquez, D.; Silva-Olivares, A.; Castro-Muñozledo, F.; Lares-Jiménez, L.F.; Rodríguez-Anaya, L.Z.; Shibayama, M.; Serrano-Luna, J. Acanthamoeba mauritaniensis genotype T4D: An environmental isolate displays pathogenic behavior. Parasitol. Int. 2020, 74, 102002. [Google Scholar] [CrossRef] [PubMed]
- Holmgaard, D.B.; Barnadas, C.; Mirbarati, S.H.; O’Brien, A.L.; Nielsen, H.V.; Stensvold, C.R. Detection and identification of acanthamoeba and other nonviral causes of infectious keratitis in corneal scrapings by real-time PCR and next-generation sequencing-based 16S–18S gene analysis. J. Clin. Microbiol. 2021, 59, e02224-20. [Google Scholar] [CrossRef]
- Megha, K.; Sharma, M.; Gupta, A.; Sehgal, R.; Khurana, S. Protein profiling of Acanthamoeba species using MALDI-TOF MS for specific identification of Acanthamoeba genotype. Parasitol. Res. 2018, 117, 729–736. [Google Scholar] [CrossRef]


| ID | Source Origin | 16S rDNA | GenBank Accession No. | % ID | Identity with Reference GenBank Accesion No. |
|---|---|---|---|---|---|
| AC11 | Corneal scraping | T4a | MF538585 | 99% | AF479525 |
| AC15 | Corneal scraping | T4a | MF538588 | 98% | AB795711 |
| AC16 | Corneal scraping | T4a | MF538586 | 98% | MK100243 |
| AC20 | Corneal scraping | T4a | MF538584 | 99% | AF479525 |
| AC22 | Corneal scraping | T4a | MF538583 | 98% | AB795713 |
| AC4 | Corneal scraping | T4b | MF563606 | 99% | AB795716 |
| AC13 | Corneal scraping | T4c | MF563608 | 94% | U03732 |
| AC25 | Corneal scraping | T4c | MF563607 | 94% | U12386 |
| AC1 | Corneal scraping | T4d | MF563605 | 99% | AF479510 |
| AC28 | Water sample | T4a | MF538587 | 98% | AB795711 |
| AC29 | Water sample | T4d | MF563604 | 99% | AF479510 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Megha, K.; Sharma, M.; Gupta, A.; Sehgal, R.; Khurana, S. Sub-Genotyping of Acanthamoeba T4 Complex: Experience from North India. Parasitologia 2023, 3, 69-78. https://doi.org/10.3390/parasitologia3010009
Megha K, Sharma M, Gupta A, Sehgal R, Khurana S. Sub-Genotyping of Acanthamoeba T4 Complex: Experience from North India. Parasitologia. 2023; 3(1):69-78. https://doi.org/10.3390/parasitologia3010009
Chicago/Turabian StyleMegha, Kirti, Megha Sharma, Amit Gupta, Rakesh Sehgal, and Sumeeta Khurana. 2023. "Sub-Genotyping of Acanthamoeba T4 Complex: Experience from North India" Parasitologia 3, no. 1: 69-78. https://doi.org/10.3390/parasitologia3010009
APA StyleMegha, K., Sharma, M., Gupta, A., Sehgal, R., & Khurana, S. (2023). Sub-Genotyping of Acanthamoeba T4 Complex: Experience from North India. Parasitologia, 3(1), 69-78. https://doi.org/10.3390/parasitologia3010009