Translocation Tales: Unraveling the MYC Deregulation in Burkitt Lymphoma for Innovative Therapeutic Strategies
Abstract
:1. Introduction
2. Epidemiology and Challenges in Treatment Approach
| Region/Country | Burkitt Lymphoma Incidence |
|---|---|
| Asia | |
| Israel | 3.77 |
| Saudi Arabia | 2.41 |
| Turkey | 2.30 |
| Republic of Korea | 1.72 |
| Japan | 1.55 |
| Jordan | 1.17 |
| Thailand | 0.88 |
| India | 0.59 |
| China | 0.45 |
| Europe | |
| Estonia | 5.67 |
| Switzerland | 4.12 |
| Belgium | 3.72 |
| Norway | 3.49 |
| Spain | 3.34 |
| Italy | 3.23 |
| The Netherlands | 3.16 |
| France | 3.13 |
| Lithuania | 3.00 |
| Denmark | 2.92 |
| United Kingdom | 2.68 |
| Ireland | 2.64 |
| Austria | 2.44 |
| Germany | 2.43 |
| Czech Republic | 1.97 |
| Belarus | 1.96 |
| Ukraine | 1.52 |
| Poland | 0.88 |
| Russian Federation | 0.72 |
| Oceania | |
| New Zealand | 3.21 |
| Australia | 3.12 |
| Africa | |
| Malawi | 19.3 |
| Uganda | 4.8 |
| Zambia | 4.2 |
| Rwanda | 3.5 |
| Burundi | 3.4 |
| South Sudan | 2.5 |
| Tanzania | 2.2 |
| Madagascar | 2.1 |
| Kenya | 1.7 |
| Mozambique | 1.7 |
| Ethiopia | 0.4 |
| Cameroon | 8.0 |
| Congo, Democratic People Republic of | 2.9 |
| Angola | 2.1 |
| Chad | 1.7 |
| Sudan | 2.0 |
| Egypt | 1.7 |
| Morocco | 1.7 |
| Algeria | 0.9 |
| South Africa | 1.6 |
| Cote d’Ivoire | 4.6 |
| Nigeria | 2.8 |
| Ghana | 2.4 |
| Senegal | 2.4 |
| Burkina Faso | 2.2 |
| Mali | 1.4 |
| Niger | 1.2 |
| Central/South America | |
| Puerto Rico | 5.32 |
| Colombia | 5.10 |
| North America | |
| US white people | 4.94 |
| United States (overall) | 4.75 |
| US black people | 4.04 |
| Canada | 2.35 |
3. MYC Deregulation in Burkitt Lymphoma: Mechanisms and Implications
- (1)
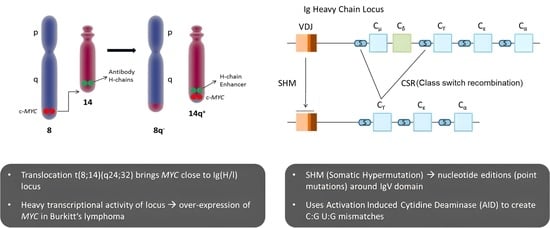
- Chromosomal Translocations: In Burkitt lymphoma (BL), the most common mechanism of MYC deregulation involves chromosomal translocations that place the MYC gene under the control of immunoglobulin (Ig) enhancer elements, leading to constitutive activation of MYC expression in B-cells [34,35,36,37]. The translocation t(8;14)(q24;q32), found in approximately 80% of BL cases, is the most frequent MYC translocation in this disease [38,39]. Additional MYC translocations in BL include t(2;8)(p12;q24) and t(8;22)(q24;q11) [39,40,41]. Constitutive MYC overexpression drives uncontrolled cell proliferation, a defining feature of BL.
- (2)
- Point Mutations: In addition to chromosomal translocations, point mutations in MYC have been identified in BL. The T58A mutation increases MYC protein stability by inhibiting its degradation via the ubiquitin–proteasome pathway and enhances MYC transcriptional activity by increasing its association with transcriptional co-activators, such as TRRAP and p300/CBP [42,43].
- (3)
- Genomic Instability: MYC translocation also contributes to the genomic instability of BL cells. MYC-induced DNA replication stress can cause DNA double-strand breaks, leading to chromosomal rearrangements and mutations [44,45]. This instability can contribute to the clonal evolution of BL and the acquisition of additional mutations that drive tumor progression and resistance to therapy [46].
- (4)
- Signaling Pathways: The molecular mechanisms of MYC-induced transformation in BL are complex and involve the deregulation of multiple signaling pathways. MYC regulates the expression of genes involved in several growth/proliferation regulating signaling pathways, including the Wnt/β-catenin, NF-κB, and PI3K/Akt/mTOR pathways [47,48]. Deregulation of these pathways can contribute to the aggressive behavior of BL and resistance to therapy.
- (5)
- MYC and Cancer Metabolism: MYC has been implicated in cancer metabolism, particularly the Warburg effect, where it promotes glucose metabolism and aerobic glycolysis in cancer cells [49,50]. This metabolic adaptation enables cancer cells to withstand nutrient scarcity and hypoxic environments. MYC also plays a role in mitochondrial biogenesis and regulation in response to growth signals and cell cycle progression [51,52,53]. This provides an opportunity for therapeutic targeting of mitochondrial factors regulated by MYC in the context of the Warburg effect in cancer.
- (6)
- MYC and Immune Regulation: Studies on human patient samples and transgenic mouse models have provided evidence that MYC is involved in the regulation of innate immune regulator CD-47 and the well-known adaptive immune checkpoint PD-L1 [54,55,56]. Saravia et. al [57] found that MYC also promotes the differentiation and activation of regulatory T-cells, which suppress immune responses and promote tumor growth. MYC influences the expression of genes involved in T-cell activation while suppressing the expression of genes involved in T-cell differentiation and function [57].
3.1. Mechanism of MYC Translocation and Its Significance in Accumulating Mutations on the Translocated Allele
3.1.1. Mechanism of IgH-MYC Translocation
3.1.2. Risk Factors for Burkitt Lymphoma and IgH-MYC Translocation
- Infection with the Epstein–Barr virus (EBV): Most people are infected with EBV at some point in their lives, but in rare cases, it can increase the risk of Burkitt lymphoma. It is believed that the virus may contribute to the occurrence of the IgH-MYC translocation [62].
- Malaria: In endemic regions (such as sub-Saharan Africa), chronic malaria infection weakens the immune system and is thought to contribute to the higher incidence of Burkitt lymphoma [63].
- Immune suppression: Individuals with weakened immune systems (such as those with HIV/AIDS or organ transplant recipients) have an increased risk [64].
3.1.3. Mutations Accumulate in the Translocated MYC Allele
3.2. Additional Genetic Alterations in Burkitt Lymphoma
3.2.1. TP53 Mutations and Its Prognostic Importance in BL
3.2.2. TCF3 and ID3 Mutations in BL
3.2.3. MYC Translocation and 11q Alterations
3.3. Evolutionary Growth Advantage as an Implication of MYC Translocation in Cancer
4. Challenges in Developing Successful Therapies against MYC-Deregulated Burkitt Lymphoma
- (1)
- Heterogeneity of MYC-deregulated BL: BL tumors display significant heterogeneity, both at the genetic and epigenetic levels, which contributes to variations in treatment response [66]. This diversity requires the development of therapies that can effectively target the range of molecular alterations in MYC-deregulated BL.
- (2)
- Tumor microenvironment: The tumor microenvironment, consisting of non-cancerous cells and extracellular matrix components, can influence tumor growth and therapy resistance in BL. The interplay between tumor cells and the microenvironment may contribute to the challenges in developing effective treatments against MYC-deregulated BL [102].
- (3)
- Off-target effects: MYC plays a crucial role in normal cell function, and its inhibition may lead to undesirable off-target effects that can cause cytotoxicity in normal cells. This further complicates the development of targeted therapies against MYC [103].
- (4)
- Complexity of MYC’s roles in cellular processes: MYC is a transcription factor that regulates a myriad of cellular processes, including cell cycle progression, metabolism, and protein synthesis [66]. This multifaceted role makes it difficult to selectively target MYC without disrupting normal cellular functions.
- (5)
- Paradoxical roles of MYC: A significantly high level of MYC can induce apoptosis or programmed cell death, as a protective mechanism against uncontrolled proliferation. This phenomenon, known as “MYC-induced apoptosis”, has been well-documented in scientific literature. For example, in a study by Evan et al. [104] it was observed that overexpression of MYC leads to both cell proliferation and apoptosis. This seemingly paradoxical effect is due to the fact that while MYC drives cell cycle progression, it also sensitizes cells to a variety of apoptotic signals. Therefore, unless a second mutation occurs that inhibits apoptosis, excess MYC can lead to cell death [104].
- (6)
- MYC’s “undruggable” nature: MYC has been considered an “undruggable” target due to the lack of well-defined small-molecule binding pockets and its intrinsically disordered structure, which hampers the development of small-molecule inhibitors [20].
- (7)
- The development of resistance: Targeted therapies often face the challenge of acquired resistance, which may emerge through the activation of alternative signaling pathways or the selection of resistant clones [105].
5. Potential Strategies to Overcome Challenges
- (1)
- Patient stratification and precision medicine: An enhanced approach to patient stratification could significantly improve therapeutic efficacy in MYC-deregulated Burkitt lymphoma. Stratification informed by molecular profiling, geographical variations (e.g., HIC vs. SSA regions), and age-related criteria can identify patient subgroups likely to benefit from targeted therapies. This holistic approach could optimize personalized treatment strategies, and help in mitigating the impact of tumor heterogeneity [106].
- (2)
- Targeting the tumor microenvironment: Therapeutic strategies that modulate the tumor microenvironment can potentially enhance the effectiveness of MYC-targeted therapies. For example, immunotherapies, such as immune checkpoint inhibitors and chimeric antigen receptor (CAR) T-cell therapy, can be combined with MYC-targeted treatments to improve antitumor immune responses [107].
- (3)
- Development of selective MYC inhibitors: The development of selective MYC inhibitors with reduced off-target effects can help in minimizing cytotoxicity in normal cells. Recent advancements in drug discovery technologies, such as structure-based drug design and high-throughput screening, can aid in identifying these selective inhibitors [108].
- (4)
- Indirect targeting of MYC: One approach to overcome MYC’s “undruggable” nature is to indirectly target it by modulating its transcription, translation, or protein stability [109]. Small molecules that inhibit MYC-MAX dimerization or target upstream signaling pathways regulating MYC have shown promise in preclinical studies [8].
- (5)
- Combination therapy: Combining targeted therapies against MYC with other treatment modalities, such as chemotherapy or immunotherapy, can potentially improve treatment outcomes and overcome resistance mechanisms [107].
6. Implications for Treatment
6.1. Background
6.2. Strategies for Targeting MYC in Burkitt Lymphoma
6.2.1. Small-Molecule Inhibitors and Targeted Therapies
6.2.2. Targeting MYC Deregulation via Cellular Pathways
- CDK9: CDK9, a component of the positive transcription elongation factor b (P-TEFb) complex, is necessary for transcriptional elongation by RNA polymerase II and is one of the prospective therapeutic targets. MYC-positive BL cells were found to be highly sensitive to CDK9 inhibition, leading to decreased cell viability and increased apoptosis [115].
- MCL-1 Inhibition: Inhibition of MCL-1, an anti-apoptotic protein that is often overexpressed in cancer cells, was found to be another potential therapeutic target for MYC-positive BL [116].
- AKT Pathway: MYC translocation activates the AKT pathway, which plays a crucial role in regulating cell survival, metabolism, and proliferation. Targeting the AKT pathway was suggested as a promising therapeutic approach for MYC-positive BL [117]. The combination of an AKT inhibitor and an mTOR inhibitor can synergistically induce apoptosis in MYC-driven lymphoma cells [117].
- Restoration of Wild-Type p53 Expression: Restoring wild-type p53 expression can inhibit their proliferation and induce apoptosis [77].
- Cell Proliferation and Survival Genes: Genes, such as CDK6, CCND2, and BCL2 could serve as potential therapeutic targets for MYC-positive BL [118]. Venetoclax, a BCL2 inhibitor, was found to be efficient in inducing apoptosis in MYC-positive lymphoma cells.
- XIAP Targeting: MYC translocation can also increase the expression of a protein known as X-linked inhibitor of apoptosis protein (XIAP), which inhibits caspase-dependent apoptosis. Targeting XIAP may increase the sensitivity of MYC-positive BL cells to apoptosis-inducing drugs [119].
- Notch and MAPK Pathways: Notch pathway was also found to be involved in MYC-driven lymphomagenesis, with MYC translocation upregulating the expression of Notch receptors and ligands, leading to the activation of the Notch pathway [120]. Therefore, targeting the mitogen-activated protein kinase (MAPK) pathway, which is active in MYC-driven malignancies, has shown therapeutic promise [121].
6.2.3. Synthetic Lethality Strategies
6.2.4. Epigenetic Modifiers
6.2.5. Leveraging MYC’s Role in Metabolism
6.2.6. Investigation of MYC Inhibitor OmoMYC
7. Challenges in Targeting MYC
8. Future Directions
9. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Epstein, M.A.; Achong, B.G.; Barr, Y.M. Virus Particles in Cultured Lymphoblasts from Burkitt’s Lymphoma. Lancet 1964, 1, 702–703. [Google Scholar] [CrossRef] [PubMed]
- Zech, L.; Haglund, U.; Nilsson, K.; Klein, G. Characteristic chromosomal abnormalities in biopsies and lymphoid-cell lines from patients with Burkitt and non-Burkitt lymphomas. Int. J. Cancer 1976, 17, 47–56. [Google Scholar] [CrossRef] [PubMed]
- Schulz, T.F.; Boshoff, C.H.; Weiss, R.A. HIV infection and neoplasia. Lancet 1996, 348, 587–591. [Google Scholar] [CrossRef] [PubMed]
- Burkitt, D.P. Etiology of Burkitt’s lymphoma—An alternative hypothesis to a vectored virus. J. Natl. Cancer Inst. 1969, 42, 19–28. [Google Scholar]
- Parkin, D.M.; Ferlay, J.; Hamdi-Chérif, M.; Sitas, F.; Thomas, J.O.; Wabinga, H.; Whelan, S.L. Cancer in Africa: Epidemiology and Prevention; IARC Press: Lyon, France, 2003. [Google Scholar]
- Orem, J.; Mbidde, E.K.; Lambert, B.; de Sanjose, S.; Weiderpass, E. Burkitt’s lymphoma in Africa, a review of the epidemiology and etiology. Afr. Health Sci. 2007, 7, 166–175. [Google Scholar]
- Chang, D.W.; Claassen, G.F.; Hann, S.R.; Cole, M.D. The c-Myc transactivation domain is a direct modulator of apoptotic versus proliferative signals. Mol. Cell. Biol. 2000, 20, 4309–4319. [Google Scholar] [CrossRef] [Green Version]
- Chen, H.; Liu, H.; Qing, G. Targeting oncogenic Myc as a strategy for cancer treatment. Signal Transduct. Target. Ther. 2018, 3, 5. [Google Scholar] [CrossRef] [Green Version]
- Dhanasekaran, R.; Deutzmann, A.; Mahauad-Fernandez, W.D.; Hansen, A.S.; Gouw, A.M.; Felsher, D.W. The MYC oncogene—The grand orchestrator of cancer growth and immune evasion. Nat. Rev. Clin. Oncol. 2022, 19, 23–36. [Google Scholar] [CrossRef]
- Kress, T.R.; Sabò, A.; Amati, B. MYC: Connecting selective transcriptional control to global RNA production. Nat. Rev. Cancer 2015, 15, 593–607. [Google Scholar] [CrossRef]
- Kalkat, M.; De Melo, J.; Hickman, K.A.; Lourenco, C.; Redel, C.; Resetca, D.; Tamachi, A.; Tu, W.B.; Penn, L.Z. MYC Deregulation in Primary Human Cancers. Genes 2017, 8, 151. [Google Scholar] [CrossRef] [Green Version]
- Beroukhim, R.; Mermel, C.H.; Porter, D.; Wei, G.; Raychaudhuri, S.; Donovan, J.; Barretina, J.; Boehm, J.S.; Dobson, J.; Urashima, M.; et al. The landscape of somatic copy-number alteration across human cancers. Nature 2010, 463, 899–905. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Boxer, L.M.; Dang, C.V. Translocations involving c-myc and c-myc function. Oncogene 2001, 20, 5595–5610. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pelengaris, S.; Khan, M. The many faces of c-MYC. Arch. Biochem. Biophys. 2003, 416, 129–136. [Google Scholar] [CrossRef] [PubMed]
- Bettess, M.D.; Dubois, N.; Murphy, M.J.; Dubey, C.; Roger, C.; Robine, S.; Trumpp, A. c-Myc is required for the formation of intestinal crypts but dispensable for homeostasis of the adult intestinal epithelium. Mol. Cell. Biol. 2005, 25, 7868–7878. [Google Scholar] [CrossRef] [Green Version]
- De la Cova, C.; Abril, M.; Bellosta, P.; Gallant, P.; Johnston, L.A. Drosophila myc regulates organ size by inducing cell competition. Cell 2004, 117, 107–116. [Google Scholar] [CrossRef] [Green Version]
- Shi, Y.; Glynn, J.M.; Guilbert, L.J.; Cotter, T.G.; Bissonnette, R.P.; Green, D.R. Role for c-myc in activation-induced apoptotic cell death in T cell hybridomas. Science 1992, 257, 212–214. [Google Scholar] [CrossRef]
- Kaczmarek, L.; Hyland, J.K.; Watt, R.; Rosenberg, M.; Baserga, R. Microinjected c-myc as a competence factor. Science 1985, 228, 1313–1315. [Google Scholar] [CrossRef]
- Johnston, J.M.; Carroll, W.L. c-myc hypermutation in Burkitt’s lymphoma. Leuk. Lymphoma 1992, 8, 431–439. [Google Scholar] [CrossRef]
- Dang, C.V. MYC on the path to cancer. Cell 2012, 149, 22–35. [Google Scholar] [CrossRef] [Green Version]
- Vecchio, E.; Fiume, G.; Correnti, S.; Romano, S.; Iaccino, E.; Mimmi, S.; Maisano, D.; Nisticò, N.; Quinto, I. Insights about MYC and Apoptosis in B-Lymphomagenesis: An Update from Murine Models. Int. J. Mol. Sci. 2020, 21, 4265. [Google Scholar] [CrossRef]
- Love, C.; Sun, Z.; Jima, D.; Li, G.; Zhang, J.; Miles, R.; Richards, K.L.; Dunphy, C.H.; Choi, W.W.; Srivastava, G.; et al. The genetic landscape of mutations in Burkitt lymphoma. Nat. Genet. 2012, 44, 1321–1325. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Burkitt, D. A sarcoma involving the jaws in African children. Br. J. Surg. 1958, 46, 218–223. [Google Scholar] [CrossRef] [PubMed]
- Gerrard, M.; Cairo, M.S.; Weston, C.; Auperin, A.; Pinkerton, R.; Lambilliote, A.; Sposto, R.; McCarthy, K.; Lacombe, M.J.; Perkins, S.L.; et al. Excellent survival following two courses of COPAD chemotherapy in children and adolescents with resected localized B-cell non-Hodgkin’s lymphoma: Results of the FAB/LMB 96 international study. Br. J. Haematol. 2008, 141, 840–847. [Google Scholar] [CrossRef] [PubMed]
- Burkhardt, B.; Oschlies, I.; Klapper, W.; Zimmermann, M.; Woessmann, W.; Meinhardt, A.; Landmann, E.; Attarbaschi, A.; Niggli, F.; Schrappe, M.; et al. Non-Hodgkin’s lymphoma in adolescents: Experiences in 378 adolescent NHL patients treated according to pediatric NHL-BFM protocols. Leukemia 2011, 25, 153–160. [Google Scholar] [CrossRef] [PubMed]
- Goldman, S.; Smith, L.; Galardy, P.; Perkins, S.L.; Frazer, J.K.; Sanger, W.; Anderson, J.R.; Gross, T.G.; Weinstein, H.; Harrison, L.; et al. Rituximab with chemotherapy in children and adolescents with central nervous system and/or bone marrow-positive Burkitt lymphoma/leukaemia: A Children’s Oncology Group Report. Br. J. Haematol. 2014, 167, 394–401. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Minard-Colin, V.; Aupérin, A.; Pillon, M.; Burke, G.A.A.; Barkauskas, D.A.; Wheatley, K.; Delgado, R.F.; Alexander, S.; Uyttebroeck, A.; Bollard, C.M.; et al. Rituximab for High-Risk, Mature B-Cell Non-Hodgkin’s Lymphoma in Children. N. Engl. J. Med. 2020, 382, 2207–2219. [Google Scholar] [CrossRef]
- Patte, C.; Auperin, A.; Michon, J.; Behrendt, H.; Leverger, G.; Frappaz, D.; Lutz, P.; Coze, C.; Perel, Y.; Raphaël, M.; et al. The Société Française d’Oncologie Pédiatrique LMB89 protocol: Highly effective multiagent chemotherapy tailored to the tumor burden and initial response in 561 unselected children with B-cell lymphomas and L3 leukemia. Blood 2001, 97, 3370–3379. [Google Scholar] [CrossRef] [Green Version]
- Ozuah, N.W.; Lubega, J.; Allen, C.E.; El-Mallawany, N.K. Five decades of low intensity and low survival: Adapting intensified regimens to cure pediatric Burkitt lymphoma in Africa. Blood Adv. 2020, 4, 4007–4019. [Google Scholar] [CrossRef]
- Mbulaiteye, S.M.; Talisuna, A.O.; Ogwang, M.D.; McKenzie, F.E.; Ziegler, J.L.; Parkin, D.M. African Burkitt’s lymphoma: Could collaboration with HIV-1 and malaria programmes reduce the high mortality rate? Lancet 2010, 375, 1661–1663. [Google Scholar] [CrossRef] [PubMed]
- Mbulaiteye, S.M.; Devesa, S.S. Burkitt Lymphoma Incidence in Five Continents. Hemato 2022, 3, 434–453. [Google Scholar] [CrossRef]
- Molyneux, E.M.; Rochford, R.; Griffin, B.; Newton, R.; Jackson, G.; Menon, G.; Harrison, C.J.; Israels, T.; Bailey, S. Burkitt’s lymphoma. Lancet 2012, 379, 1234–1244. [Google Scholar] [CrossRef] [Green Version]
- Bornkamm, G.W. Epstein-Barr virus and its role in the pathogenesis of Burkitt’s lymphoma: An unresolved issue. Semin. Cancer Biol. 2009, 19, 351–365. [Google Scholar] [CrossRef] [PubMed]
- Dalla-Favera, R.; Bregni, M.; Erikson, J.; Patterson, D.; Gallo, R.C.; Croce, C.M. Human c-myc onc gene is located on the region of chromosome 8 that is translocated in Burkitt lymphoma cells. Proc. Natl. Acad. Sci. USA 1982, 79, 7824–7827. [Google Scholar] [CrossRef]
- Taub, R.; Kirsch, I.; Morton, C.; Lenoir, G.; Swan, D.; Tronick, S.; Aaronson, S.; Leder, P. Translocation of the c-myc gene into the immunoglobulin heavy chain locus in human Burkitt lymphoma and murine plasmacytoma cells. Proc. Natl. Acad. Sci. USA 1982, 79, 7837–7841. [Google Scholar] [CrossRef] [PubMed]
- Hamlyn, P.H.; Rabbitts, T.H. Translocation joins c-myc and immunoglobulin gamma 1 genes in a Burkitt lymphoma revealing a third exon in the c-myc oncogene. Nature 1983, 304, 135–139. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, L.; Papenhausen, P.; Shao, H. The Role of c-MYC in B-Cell Lymphomas: Diagnostic and Molecular Aspects. Genes 2017, 8, 116. [Google Scholar] [CrossRef] [Green Version]
- Basso, K.; Frascella, E.; Zanesco, L.; Rosolen, A. Improved long-distance polymerase chain reaction for the detection of t(8;14)(q24;q32) in Burkitt’s lymphomas. Am. J. Pathol. 1999, 155, 1479–1485. [Google Scholar] [CrossRef] [PubMed]
- Blum, K.A.; Lozanski, G.; Byrd, J.C. Adult Burkitt leukemia and lymphoma. Blood 2004, 104, 3009–3020. [Google Scholar] [CrossRef]
- Hecht, J.L.; Aster, J.C. Molecular biology of Burkitt’s lymphoma. J. Clin. Oncol. 2000, 18, 3707–3721. [Google Scholar] [CrossRef] [PubMed]
- Neri, A.; Barriga, F.; Knowles, D.M.; Magrath, I.T.; Dalla-Favera, R. Different regions of the immunoglobulin heavy-chain locus are involved in chromosomal translocations in distinct pathogenetic forms of Burkitt lymphoma. Proc. Natl. Acad. Sci. USA 1988, 85, 2748–2752. [Google Scholar] [CrossRef] [PubMed]
- Devaiah, B.N.; Mu, J.; Akman, B.; Uppal, S.; Weissman, J.D.; Cheng, D.; Baranello, L.; Nie, Z.; Levens, D.; Singer, D.S. MYC protein stability is negatively regulated by BRD4. Proc. Natl. Acad. Sci. USA 2020, 117, 13457–13467. [Google Scholar] [CrossRef] [PubMed]
- Hinds, J.W.; Feris, E.J.; Wilkins, O.M.; Deary, L.T.; Wang, X.; Cole, M.D. S146L in MYC is a context-dependent activating substitution in cancer development. PLoS ONE 2022, 17, e0272771. [Google Scholar] [CrossRef] [PubMed]
- Kuzyk, A.; Mai, S. c-MYC-induced genomic instability. Cold Spring Harb. Perspect. Med. 2014, 4, a014373. [Google Scholar] [CrossRef]
- Kumari, A.; Folk, W.P.; Sakamuro, D. The Dual Roles of MYC in Genomic Instability and Cancer Chemoresistance. Genes 2017, 8, 158. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Curti, L.; Campaner, S. MYC-Induced Replicative Stress: A Double-Edged Sword for Cancer Development and Treatment. Int. J. Mol. Sci. 2021, 22, 6168. [Google Scholar] [CrossRef]
- Shortt, J.; Martin, B.P.; Newbold, A.; Hannan, K.M.; Devlin, J.R.; Baker, A.J.; Ralli, R.; Cullinane, C.; Schmitt, C.A.; Reimann, M.; et al. Combined inhibition of PI3K-related DNA damage response kinases and mTORC1 induces apoptosis in MYC-driven B-cell lymphomas. Blood 2013, 121, 2964–2974. [Google Scholar] [CrossRef] [Green Version]
- Han, S.S.; Yun, H.; Son, D.J.; Tompkins, V.S.; Peng, L.; Chung, S.T.; Kim, J.S.; Park, E.S.; Janz, S. NF-kappaB/STAT3/PI3K signaling crosstalk in iMyc E mu B lymphoma. Mol. Cancer 2010, 9, 97. [Google Scholar] [CrossRef] [Green Version]
- Dang, C.V. The interplay between MYC and HIF in the Warburg effect. In Ernst Schering Foundation Symposium Proceedings; Springer: Berlin/Heidelberg, Germany, 2007; pp. 35–53. [Google Scholar]
- Dang, C.V. MYC, metabolism, cell growth, and tumorigenesis. Cold Spring Harb. Perspect. Med. 2013, 3, a014217. [Google Scholar] [CrossRef] [Green Version]
- Kim, J.; Lee, J.H.; Iyer, V.R. Global identification of Myc target genes reveals its direct role in mitochondrial biogenesis and its E-box usage in vivo. PLoS ONE 2008, 3, e1798. [Google Scholar] [CrossRef] [Green Version]
- Popay, T.M.; Wang, J.; Adams, C.M.; Howard, G.C.; Codreanu, S.G.; Sherrod, S.D.; McLean, J.A.; Thomas, L.R.; Lorey, S.L.; Machida, Y.J.; et al. MYC regulates ribosome biogenesis and mitochondrial gene expression programs through its interaction with host cell factor-1. eLife 2021, 10, e60191. [Google Scholar] [CrossRef]
- Morrish, F.; Hockenbery, D. MYC and mitochondrial biogenesis. Cold Spring Harb. Perspect. Med. 2014, 4, a014225. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Casey, S.C.; Baylot, V.; Felsher, D.W. The MYC oncogene is a global regulator of the immune response. Blood 2018, 131, 2007–2015. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Casey, S.C.; Tong, L.; Li, Y.; Do, R.; Walz, S.; Fitzgerald, K.N.; Gouw, A.M.; Baylot, V.; Gütgemann, I.; Eilers, M.; et al. MYC regulates the antitumor immune response through CD47 and PD-L1. Science 2016, 352, 227–231. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, E.Y.; Kim, A.; Kim, S.K.; Chang, Y.S. MYC expression correlates with PD-L1 expression in non-small cell lung cancer. Lung Cancer 2017, 110, 63–67. [Google Scholar] [CrossRef]
- Saravia, J.; Zeng, H.; Dhungana, Y.; Bastardo Blanco, D.; Nguyen, T.M.; Chapman, N.M.; Wang, Y.; Kanneganti, A.; Liu, S.; Raynor, J.L.; et al. Homeostasis and transitional activation of regulatory T cells require c-Myc. Sci. Adv. 2020, 6, eaaw6443. [Google Scholar] [CrossRef] [Green Version]
- Cerami, E.; Gao, J.; Dogrusoz, U.; Gross, B.E.; Sumer, S.O.; Aksoy, B.A.; Jacobsen, A.; Byrne, C.J.; Heuer, M.L.; Larsson, E.; et al. The cBio Cancer Genomics Portal: An Open Platform for Exploring Multidimensional Cancer Genomics Data. Cancer Discov. 2012, 2, 401. [Google Scholar] [CrossRef] [Green Version]
- Gao, J.; Aksoy, B.A.; Dogrusoz, U.; Dresdner, G.; Gross, B.; Sumer, S.O.; Sun, Y.; Jacobsen, A.; Sinha, R.; Larsson, E.; et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci. Signal. 2013, 6, pl1. [Google Scholar] [CrossRef] [Green Version]
- Meyer, N.; Penn, L.Z. Reflecting on 25 years with MYC. Nat. Rev. Cancer 2008, 8, 976–990. [Google Scholar] [CrossRef]
- Stasevich, E.M.; Uvarova, A.N.; Murashko, M.M.; Khabusheva, E.R.; Sheetikov, S.A.; Prassolov, V.S.; Kuprash, D.V.; Demin, D.E.; Schwartz, A.M. Enhancer RNA AL928768.3 from the IGH Locus Regulates MYC Expression and Controls the Proliferation and Chemoresistance of Burkitt Lymphoma Cells with IGH/MYC Translocation. Int. J. Mol. Sci. 2022, 23, 4624. [Google Scholar] [CrossRef]
- Brady, G.; MacArthur, G.J.; Farrell, P.J. Epstein-Barr virus and Burkitt lymphoma. J. Clin. Pathol. 2007, 60, 1397–1402. [Google Scholar] [CrossRef] [Green Version]
- Broen, K.; Dickens, J.; Trangucci, R.; Ogwang, M.D.; Tenge, C.N.; Masalu, N.; Reynolds, S.J.; Kawira, E.; Kerchan, P.; Were, P.A.; et al. Burkitt lymphoma risk shows geographic and temporal associations with Plasmodium falciparum infections in Uganda, Tanzania, and Kenya. Proc. Natl. Acad. Sci. USA 2023, 120, e2211055120. [Google Scholar] [CrossRef] [PubMed]
- Blinder, V.S.; Chadburn, A.; Furman, R.R.; Mathew, S.; Leonard, J.P. Improving outcomes for patients with Burkitt lymphoma and HIV. AIDS Patient Care STDs 2008, 22, 175–187. [Google Scholar] [CrossRef] [PubMed]
- Bemark, M.; Neuberger, M.S. The c-MYC allele that is translocated into the IgH locus undergoes constitutive hypermutation in a Burkitt’s lymphoma line. Oncogene 2000, 19, 3404–3410. [Google Scholar] [CrossRef] [Green Version]
- Schmitz, R.; Ceribelli, M.; Pittaluga, S.; Wright, G.; Staudt, L.M. Oncogenic mechanisms in Burkitt lymphoma. Cold Spring Harb. Perspect. Med. 2014, 4, a014282. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hummel, M.; Bentink, S.; Berger, H.; Klapper, W.; Wessendorf, S.; Barth, T.F.; Bernd, H.W.; Cogliatti, S.B.; Dierlamm, J.; Feller, A.C.; et al. A biologic definition of Burkitt’s lymphoma from transcriptional and genomic profiling. N. Engl. J. Med. 2006, 354, 2419–2430. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- López, C.; Kleinheinz, K.; Aukema, S.M.; Rohde, M.; Bernhart, S.H.; Hübschmann, D.; Wagener, R.; Toprak, U.H.; Raimondi, F.; Kreuz, M.; et al. Genomic and transcriptomic changes complement each other in the pathogenesis of sporadic Burkitt lymphoma. Nat. Commun. 2019, 10, 1459. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Küppers, R.; Dalla-Favera, R. Mechanisms of chromosomal translocations in B cell lymphomas. Oncogene 2001, 20, 5580–5594. [Google Scholar] [CrossRef] [Green Version]
- Tandon, A.; Birkenhagen, J.; Nagalla, D.; Kölker, S.; Sauer, S.W. ADP-dependent glucokinase as a novel onco-target for haematological malignancies. Sci. Rep. 2020, 10, 13584. [Google Scholar] [CrossRef]
- Schmitz, R.; Young, R.M.; Ceribelli, M.; Jhavar, S.; Xiao, W.; Zhang, M.; Wright, G.; Shaffer, A.L.; Hodson, D.J.; Buras, E.; et al. Burkitt lymphoma pathogenesis and therapeutic targets from structural and functional genomics. Nature 2012, 490, 116–120. [Google Scholar] [CrossRef] [Green Version]
- Sánchez-Beato, M.; Sánchez-Aguilera, A.; Piris, M.A. Cell cycle deregulation in B-cell lymphomas. Blood 2003, 101, 1220–1235. [Google Scholar] [CrossRef] [Green Version]
- Newman, A.M.; Zaka, M.; Zhou, P.; Blain, A.E.; Erhorn, A.; Barnard, A.; Crossland, R.E.; Wilkinson, S.; Enshaei, A.; De Zordi, J.; et al. Genomic abnormalities of TP53 define distinct risk groups of paediatric B-cell non-Hodgkin lymphoma. Leukemia 2022, 36, 781–789. [Google Scholar] [CrossRef] [PubMed]
- López, C.; Burkhardt, B.; Chan, J.K.C.; Leoncini, L.; Mbulaiteye, S.M.; Ogwang, M.D.; Orem, J.; Rochford, R.; Roschewski, M.; Siebert, R. Burkitt lymphoma. Nat. Rev. Dis. Prim. 2022, 8, 78. [Google Scholar] [CrossRef] [PubMed]
- Leventaki, V.; Rodic, V.; Tripp, S.R.; Bayerl, M.G.; Perkins, S.L.; Barnette, P.; Schiffman, J.D.; Miles, R.R. TP53 pathway analysis in paediatric Burkitt lymphoma reveals increased MDM4 expression as the only TP53 pathway abnormality detected in a subset of cases. Br. J. Haematol. 2012, 158, 763–771. [Google Scholar] [CrossRef] [PubMed]
- Burkhardt, B.; Michgehl, U.; Rohde, J.; Erdmann, T.; Berning, P.; Reutter, K.; Rohde, M.; Borkhardt, A.; Burmeister, T.; Dave, S.; et al. Clinical relevance of molecular characteristics in Burkitt lymphoma differs according to age. Nat. Commun. 2022, 13, 3881. [Google Scholar] [CrossRef] [PubMed]
- Yu, D.; Carroll, M.; Thomas-Tikhonenko, A. p53 status dictates responses of B lymphomas to monotherapy with proteasome inhibitors. Blood 2007, 109, 4936–4943. [Google Scholar] [CrossRef] [PubMed]
- Richter, J.; Schlesner, M.; Hoffmann, S.; Kreuz, M.; Leich, E.; Burkhardt, B.; Rosolowski, M.; Ammerpohl, O.; Wagener, R.; Bernhart, S.H.; et al. Recurrent mutation of the ID3 gene in Burkitt lymphoma identified by integrated genome, exome and transcriptome sequencing. Nat. Genet. 2012, 44, 1316–1320. [Google Scholar] [CrossRef] [PubMed]
- Rohde, M.; Bonn, B.R.; Zimmermann, M.; Lange, J.; Möricke, A.; Klapper, W.; Oschlies, I.; Szczepanowski, M.; Nagel, I.; Schrappe, M.; et al. Relevance of ID3-TCF3-CCND3 pathway mutations in pediatric aggressive B-cell lymphoma treated according to the non-Hodgkin Lymphoma Berlin-Frankfurt-Münster protocols. Haematologica 2017, 102, 1091–1098. [Google Scholar] [CrossRef] [Green Version]
- Swerdlow, S.H.; Campo, E.; Pileri, S.A.; Harris, N.L.; Stein, H.; Siebert, R.; Advani, R.; Ghielmini, M.; Salles, G.A.; Zelenetz, A.D.; et al. The 2016 revision of the World Health Organization classification of lymphoid neoplasms. Blood 2016, 127, 2375–2390. [Google Scholar] [CrossRef] [Green Version]
- Lin, P.; Dickason, T.J.; Fayad, L.E.; Lennon, P.A.; Hu, P.; Garcia, M.; Routbort, M.J.; Miranda, R.; Wang, X.; Qiao, W.; et al. Prognostic value of MYC rearrangement in cases of B-cell lymphoma, unclassifiable, with features intermediate between diffuse large B-cell lymphoma and Burkitt lymphoma. Cancer 2012, 118, 1566–1573. [Google Scholar] [CrossRef]
- Salaverria, I.; Martin-Guerrero, I.; Wagener, R.; Kreuz, M.; Kohler, C.W.; Richter, J.; Pienkowska-Grela, B.; Adam, P.; Burkhardt, B.; Claviez, A.; et al. A recurrent 11q aberration pattern characterizes a subset of MYC-negative high-grade B-cell lymphomas resembling Burkitt lymphoma. Blood 2014, 123, 1187–1198. [Google Scholar] [CrossRef] [Green Version]
- García-Gutiérrez, L.; Delgado, M.D.; León, J. MYC Oncogene Contributions to Release of Cell Cycle Brakes. Genes 2019, 10, 244. [Google Scholar] [CrossRef] [Green Version]
- Mlynarczyk, C.; Fontán, L.; Melnick, A. Germinal center-derived lymphomas: The darkest side of humoral immunity. Immunol. Rev. 2019, 288, 214–239. [Google Scholar] [CrossRef] [Green Version]
- Lindström, M.S.; Wiman, K.G. Role of genetic and epigenetic changes in Burkitt lymphoma. Semin. Cancer Biol. 2002, 12, 381–387. [Google Scholar] [CrossRef] [PubMed]
- McMahon, S.B.; Wood, M.A.; Cole, M.D. The essential cofactor TRRAP recruits the histone acetyltransferase hGCN5 to c-Myc. Mol. Cell. Biol. 2000, 20, 556–562. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Park, J.; Kunjibettu, S.; McMahon, S.B.; Cole, M.D. The ATM-related domain of TRRAP is required for histone acetyltransferase recruitment and Myc-dependent oncogenesis. Genes Dev. 2001, 15, 1619–1624. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ribeiro, M.L.; Reyes-Garau, D.; Armengol, M.; Fernández-Serrano, M.; Roué, G. Recent Advances in the Targeting of Epigenetic Regulators in B-Cell Non-Hodgkin Lymphoma. Front. Genet. 2019, 10, 986. [Google Scholar] [CrossRef] [PubMed]
- Fernández-Serrano, M.; Winkler, R.; Santos, J.C.; Le Pannérer, M.M.; Buschbeck, M.; Roué, G. Histone Modifications and Their Targeting in Lymphoid Malignancies. Int. J. Mol. Sci. 2021, 23, 253. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Van Calcar, S.; Qu, C.; Cavenee, W.K.; Zhang, M.Q.; Ren, B. A global transcriptional regulatory role for c-Myc in Burkitt’s lymphoma cells. Proc. Natl. Acad. Sci. USA 2003, 100, 8164–8169. [Google Scholar] [CrossRef] [PubMed]
- Cowling, V.H.; Turner, S.A.; Cole, M.D. Burkitt’s lymphoma-associated c-Myc mutations converge on a dramatically altered target gene response and implicate Nol5a/Nop56 in oncogenesis. Oncogene 2014, 33, 3519–3527. [Google Scholar] [CrossRef] [Green Version]
- Poole, C.J.; Zheng, W.; Lodh, A.; Yevtodiyenko, A.; Liefwalker, D.; Li, H.; Felsher, D.W.; van Riggelen, J. DNMT3B overexpression contributes to aberrant DNA methylation and MYC-driven tumor maintenance in T-ALL and Burkitt’s lymphoma. Oncotarget 2017, 8, 76898–76920. [Google Scholar] [CrossRef] [Green Version]
- Videtta, A.D.; Malagnino, V.; De Falco, G. Current understanding of the role and regulation of miRNAs in Burkitt lymphoma. Blood Lymphat. Cancer Targets Ther. 2018, 8, 33–45. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shams, R.; Asadzadeh Aghdaei, H.; Behmanesh, A.; Sadeghi, A.; Zali, M.; Salari, S.; Padrón, J.M. MicroRNAs Targeting MYC Expression: Trace of Hope for Pancreatic Cancer Therapy. A Systematic Review. Cancer Manag. Res. 2020, 12, 2393–2404. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Onnis, A.; De Falco, G.; Antonicelli, G.; Onorati, M.; Bellan, C.; Sherman, O.; Sayed, S.; Leoncini, L. Alteration of microRNAs regulated by c-Myc in Burkitt lymphoma. PLoS ONE 2010, 5, e12960. [Google Scholar] [CrossRef] [PubMed]
- Bui, T.V.; Mendell, J.T. Myc: Maestro of microRNAs. Genes Cancer 2010, 1, 568–575. [Google Scholar] [CrossRef]
- Oduor, C.I.; Kaymaz, Y.; Chelimo, K.; Otieno, J.A.; Ong’echa, J.M.; Moormann, A.M.; Bailey, J.A. Integrative microRNA and mRNA deep-sequencing expression profiling in endemic Burkitt lymphoma. BMC Cancer 2017, 17, 761. [Google Scholar] [CrossRef] [Green Version]
- Klanova, M.; Klener, P. BCL-2 Proteins in Pathogenesis and Therapy of B-Cell Non-Hodgkin Lymphomas. Cancers 2020, 12, 938. [Google Scholar] [CrossRef] [Green Version]
- Meškytė, E.M.; Keskas, S.; Ciribilli, Y. MYC as a Multifaceted Regulator of Tumor Microenvironment Leading to Metastasis. Int. J. Mol. Sci. 2020, 21, 7710. [Google Scholar] [CrossRef]
- Dews, M.; Homayouni, A.; Yu, D.; Murphy, D.; Sevignani, C.; Wentzel, E.; Furth, E.E.; Lee, W.M.; Enders, G.H.; Mendell, J.T.; et al. Augmentation of tumor angiogenesis by a Myc-activated microRNA cluster. Nat. Genet. 2006, 38, 1060–1065. [Google Scholar] [CrossRef] [Green Version]
- Whitfield, J.R.; Soucek, L. Tumor microenvironment: Becoming sick of Myc. Cell. Mol. Life Sci. 2012, 69, 931–934. [Google Scholar] [CrossRef] [Green Version]
- Balkwill, F.R.; Capasso, M.; Hagemann, T. The tumor microenvironment at a glance. J. Cell Sci. 2012, 125 Pt 23, 5591–5596. [Google Scholar] [CrossRef] [Green Version]
- Soucek, L.; Whitfield, J.; Martins, C.P.; Finch, A.J.; Murphy, D.J.; Sodir, N.M.; Karnezis, A.N.; Swigart, L.B.; Nasi, S.; Evan, G.I. Modelling Myc inhibition as a cancer therapy. Nature 2008, 455, 679–683. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Evan, G.I.; Wyllie, A.H.; Gilbert, C.S.; Littlewood, T.D.; Land, H.; Brooks, M.; Waters, C.M.; Penn, L.Z.; Hancock, D.C. Induction of apoptosis in fibroblasts by c-myc protein. Cell 1992, 69, 119–128. [Google Scholar] [CrossRef] [PubMed]
- Sabnis, A.J.; Bivona, T.G. Principles of Resistance to Targeted Cancer Therapy: Lessons from Basic and Translational Cancer Biology. Trends Mol. Med. 2019, 25, 185–197. [Google Scholar] [CrossRef] [PubMed]
- Gerlinger, M.; Rowan, A.J.; Horswell, S.; Math, M.; Larkin, J.; Endesfelder, D.; Gronroos, E.; Martinez, P.; Matthews, N.; Stewart, A.; et al. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N. Engl. J. Med. 2012, 366, 883–892. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Armand, P. Immune checkpoint blockade in hematologic malignancies. Blood 2015, 125, 3393–3400. [Google Scholar] [CrossRef] [PubMed]
- Koh, C.M.; Sabò, A.; Guccione, E. Targeting MYC in cancer therapy: RNA processing offers new opportunities. BioEssays News Rev. Mol. Cell. Dev. Biol. 2016, 38, 266–275. [Google Scholar] [CrossRef] [Green Version]
- Delmore, J.E.; Issa, G.C.; Lemieux, M.E.; Rahl, P.B.; Shi, J.; Jacobs, H.M.; Kastritis, E.; Gilpatrick, T.; Paranal, R.M.; Qi, J.; et al. BET bromodomain inhibition as a therapeutic strategy to target c-Myc. Cell 2011, 146, 904–917. [Google Scholar] [CrossRef] [Green Version]
- Pagano, L.; Caira, M.; Valentini, C.G.; Fianchi, L. Clinical aspects and therapy of sporadic burkitt lymphoma. Mediterr. J. Hematol. Infect. Dis. 2009, 1, e2009030. [Google Scholar] [CrossRef]
- Thomas, D.A.; Faderl, S.; O’Brien, S.; Bueso-Ramos, C.; Cortes, J.; Garcia-Manero, G.; Giles, F.J.; Verstovsek, S.; Wierda, W.G.; Pierce, S.A.; et al. Chemoimmunotherapy with hyper-CVAD plus rituximab for the treatment of adult Burkitt and Burkitt-type lymphoma or acute lymphoblastic leukemia. Cancer 2006, 106, 1569–1580. [Google Scholar] [CrossRef]
- Llombart, V.; Mansour, M.R. Therapeutic targeting of “undruggable” MYC. eBioMedicine 2022, 75, 103756. [Google Scholar] [CrossRef]
- Tang, M.; O’Grady, S.; Crown, J.; Duffy, M.J. MYC as a therapeutic target for the treatment of triple-negative breast cancer: Preclinical investigations with the novel MYC inhibitor, MYCi975. Breast Cancer Res. Treat. 2022, 195, 105–115. [Google Scholar] [CrossRef]
- Huang, M.J.; Cheng, Y.C.; Liu, C.R.; Lin, S.; Liu, H.E. A small-molecule c-Myc inhibitor, 10058-F4, induces cell-cycle arrest, apoptosis, and myeloid differentiation of human acute myeloid leukemia. Exp. Hematol. 2006, 34, 1480–1489. [Google Scholar] [CrossRef] [PubMed]
- Sakamoto, T.; Ajiro, M.; Watanabe, A.; Matsushima, S.; Ueda, K.; Hagiwara, M. Application of the CDK9 inhibitor FIT-039 for the treatment of KSHV-associated malignancy. BMC Cancer 2023, 23, 71. [Google Scholar] [CrossRef] [PubMed]
- Daly, B.T.; Ippolito, M.T.; Gu, J.J.; Mavis, M.C.; Torka, P.; Hernandez-Ilizaliturri, F.J.; Barth, M.J. MCL-1 Inhibition by the Selective MCL-1 Inhibitor AMG-176 Induces in Vitro Activity Against Burkitt Lymphoma Cell Lines and Synergistically Enhances the Cytotoxic Effect of Chemotherapy and BH3 Mimetics. Blood 2019, 134 (Suppl. 1), 5303. [Google Scholar] [CrossRef]
- Ahmadi, S.E.; Rahimi, S.; Zarandi, B.; Chegeni, R.; Safa, M. MYC: A multipurpose oncogene with prognostic and therapeutic implications in blood malignancies. J. Hematol. Oncol. 2021, 14, 121. [Google Scholar] [CrossRef] [PubMed]
- Sun, R.; Medeiros, L.J.; Young, K.H. Diagnostic and predictive biomarkers for lymphoma diagnosis and treatment in the era of precision medicine. Mod. Pathol. 2016, 29, 1118–1142. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Satta, T.; Grant, S. Enhancing venetoclax activity in hematological malignancies. Expert Opin. Investig. Drugs 2020, 29, 697–708. [Google Scholar] [CrossRef]
- Pozzo, F.; Bittolo, T.; Vendramini, E.; Bomben, R.; Bulian, P.; Rossi, F.M.; Zucchetto, A.; Tissino, E.; Degan, M.; D’Arena, G.; et al. NOTCH1-mutated chronic lymphocytic leukemia cells are characterized by a MYC-related overexpression of nucleophosmin 1 and ribosome-associated components. Leukemia 2017, 31, 2407–2415. [Google Scholar] [CrossRef]
- Lovec, H.; Grzeschiczek, A.; Kowalski, M.B.; Möröy, T. Cyclin D1/bcl-1 cooperates with myc genes in the generation of B-cell lymphoma in transgenic mice. EMBO J. 1994, 13, 3487–3495. [Google Scholar] [CrossRef]
- Thng, D.K.H.; Toh, T.B.; Chow, E.K. Capitalizing on Synthetic Lethality of MYC to Treat Cancer in the Digital Age. Trends Pharmacol. Sci. 2021, 42, 166–182. [Google Scholar] [CrossRef]
- Zhang, J.; Zhang, S.; Shi, Q.; Yang, D.; Allen, T.D. Pro-Survival Bcl-2 Proteins Are Modifiers of MYC-VX-680 Synthetic Lethality. J. Cell. Immunol. 2020, 2, 227–232. [Google Scholar] [CrossRef]
- Mertz, J.A.; Conery, A.R.; Bryant, B.M.; Sandy, P.; Balasubramanian, S.; Mele, D.A.; Bergeron, L.; Sims, R.J. Targeting MYC dependence in cancer by inhibiting BET bromodomains. Proc. Natl. Acad. Sci. USA 2011, 108, 16669–16674. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Li, D.; Zhou, J. Histone Deacetylase 6 as a Therapeutic Target in B Cell-Associated Hematological Malignancies. Front. Pharmacol. 2020, 11, 971. [Google Scholar] [CrossRef] [PubMed]
- Ma, Z.-G.; Kong, C.-Y.; Wu, H.-M.; Song, P.; Zhang, X.; Yuan, Y.-P.; Deng, W.; Tang, Q.-Z. Toll-like Receptor 5 Deficiency Diminishes Doxorubicin-Induced Acute Cardiotoxicity in Mice. Theranostics 2020, 10, 11013–11025. [Google Scholar] [CrossRef]
- Yuneva, M.; Zamboni, N.; Oefner, P.; Sachidanandam, R.; Lazebnik, Y. Deficiency in glutamine but not glucose induces MYC-dependent apoptosis in human cells. J. Cell Biol. 2007, 178, 93–105. [Google Scholar] [CrossRef] [Green Version]
- Gao, P.; Tchernyshyov, I.; Chang, T.-C.; Lee, Y.-S.; Kita, K.; Ochi, T.; Zeller, K.I.; De Marzo, A.M.; Van Eyk, J.E.; Mendell, J.T.; et al. c-Myc suppression of miR-23a/b enhances mitochondrial glutaminase expression and glutamine metabolism. Nature 2009, 458, 762–765. [Google Scholar] [CrossRef] [Green Version]
- Mullen, A.R.; Wheaton, W.W.; Jin, E.S.; Chen, P.-H.; Sullivan, L.B.; Cheng, T.; Yang, Y.; Linehan, W.M.; Chandel, N.S.; DeBerardinis, R.J. Reductive carboxylation supports growth in tumour cells with defective mitochondria. Nature 2011, 481, 385–388. [Google Scholar] [CrossRef] [Green Version]
- Metallo, C.M.; Gameiro, P.A.; Bell, E.L.; Mattaini, K.R.; Yang, J.; Hiller, K.; Jewell, C.M.; Johnson, Z.R.; Irvine, D.J.; Guarente, L.; et al. Reductive glutamine metabolism by IDH1 mediates lipogenesis under hypoxia. Nature 2011, 481, 380–384. [Google Scholar] [CrossRef] [Green Version]
- Jacque, N.; Ronchetti, A.M.; Larrue, C.; Meunier, G.; Birsen, R.; Willems, L.; Saland, E.; Decroocq, J.; Maciel, T.T.; Lambert, M.; et al. Targeting glutaminolysis has antileukemic activity in acute myeloid leukemia and synergizes with BCL-2 inhibition. Blood 2015, 126, 1346–1356. [Google Scholar] [CrossRef] [Green Version]
- Savino, M.; Annibali, D.; Carucci, N.; Favuzzi, E.; Cole, M.D.; Evan, G.I.; Soucek, L.; Nasi, S. The action mechanism of the Myc inhibitor termed Omomyc may give clues on how to target Myc for cancer therapy. PLoS ONE 2011, 6, e22284. [Google Scholar] [CrossRef] [Green Version]
- Jung, L.A.; Gebhardt, A.; Koelmel, W.; Ade, C.P.; Walz, S.; Kuper, J.; von Eyss, B.; Letschert, S.; Redel, C.; d’Artista, L.; et al. OmoMYC blunts promoter invasion by oncogenic MYC to inhibit gene expression characteristic of MYC-dependent tumors. Oncogene 2017, 36, 1911–1924. [Google Scholar] [CrossRef] [PubMed] [Green Version]



| Mechanism | Description |
|---|---|
| Gene Amplification | This is when there is an increase in the number of copies of the MYC gene, leading to overproduction of MYC protein. |
| Chromosomal Translocation | This is when the MYC gene is moved to a different chromosome, often leading to its inappropriate activation. In Burkitt lymphoma, for example, MYC is often translocated to the immunoglobulin heavy or light chain loci, which are highly active regions of the genome. |
| Mutation | Mutations in the MYC gene or in the genes that regulate MYC can lead to increased MYC activity. |
| Deregulation of Transcriptional Control | Disruptions in the normal mechanisms that control the transcription of the MYC gene can lead to overproduction of the MYC protein. |
| Post-transcriptional and Post-translational Modifications | Changes that occur to MYC mRNA or MYC protein after transcription and translation, respectively, can increase the stability, abundance, or activity of MYC. |
| Deregulation of MYC Degradation | Normally, MYC protein is rapidly degraded to maintain proper control of its levels. Disruptions in these degradation pathways can lead to increased levels of MYC. |
| Mechanism of Action | Type | Drugs |
|---|---|---|
| Anti-MYC (MYC Proto-Oncogene Protein) | Protein | 6 |
| Aurora kinase A (AURKA; ARK1)/c-MYC Interaction Inhibitors | Protein | 4 |
| Drugs Targeting MYC Proto-Oncogene Protein Promoter G-quadruplex (MycG4) | Gene | 8 |
| Drugs Targeting MYC Transcription Factors | Protein | 12 |
| Dual Specificity Mitogen-Activated Protein Kinase 3 (MAP2K3)/c-MYC Interaction Inhibitors | Protein | 1 |
| MYC Expression Inhibitors | Gene | 152 |
| MYC Proto-Oncogene Protein (c-MYC) Degradation Inducers | Protein | 46 |
| MYC Proto-Oncogene Protein (c-MYC) Gene Editing | Gene | 1 |
| MYC Proto-Oncogene Protein (c-MYC) Inhibitors | Protein | 159 |
| MYC Proto-Oncogene Protein (c-MYC)/MYC-Associated Factor X (Max) Interaction Inhibitors | Protein | 24 |
| MYC Proto-Oncogene Protein (MYC)/TRRAP Interaction Inhibitors | Protein | 2 |
| Transcription Factor Ligands | Protein | 35 |
| WD Repeat-Containing Protein 5 (WDR5; BIG3)/c-MYC Interaction Inhibitors | Protein | 8 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tandon, A.; Kuriappan, J.A.; Dubey, V. Translocation Tales: Unraveling the MYC Deregulation in Burkitt Lymphoma for Innovative Therapeutic Strategies. Lymphatics 2023, 1, 97-117. https://doi.org/10.3390/lymphatics1020010
Tandon A, Kuriappan JA, Dubey V. Translocation Tales: Unraveling the MYC Deregulation in Burkitt Lymphoma for Innovative Therapeutic Strategies. Lymphatics. 2023; 1(2):97-117. https://doi.org/10.3390/lymphatics1020010
Chicago/Turabian StyleTandon, Amol, Jissy Akkarapattiakal Kuriappan, and Vaibhav Dubey. 2023. "Translocation Tales: Unraveling the MYC Deregulation in Burkitt Lymphoma for Innovative Therapeutic Strategies" Lymphatics 1, no. 2: 97-117. https://doi.org/10.3390/lymphatics1020010
APA StyleTandon, A., Kuriappan, J. A., & Dubey, V. (2023). Translocation Tales: Unraveling the MYC Deregulation in Burkitt Lymphoma for Innovative Therapeutic Strategies. Lymphatics, 1(2), 97-117. https://doi.org/10.3390/lymphatics1020010