Epigenetics of Melanoma
A topical collection in Epigenomes (ISSN 2075-4655).
Viewed by 15041
Share This Topical Collection
Editor
 Dr. Ivana De la Serna
Dr. Ivana De la Serna
 Dr. Ivana De la Serna
Dr. Ivana De la Serna
E-Mail
Website
Collection Editor
Cancer Biology, College of Medicine and Life Sciences, University of Toledo, Toledo, OH 43614, USA
Interests: SWI/SNF chromatin remodeling enzymes; melanocyte differentiation and regulation of pigmentation; epigenetic regulation of the response to ultraviolet radiation; cancer epigenetics
Special Issues, Collections and Topics in MDPI journals
Topical Collection Information
Dear Colleagues,
Melanoma is an aggressive skin cancer that readily metastasizes and is resistant to conventional therapies. Newer treatments which target a common mutation in the BRAF gene as well as immunotherapy have been more effective than conventional therapies, but still not curative for all patients due to innate and acquired resistance mechanisms. The melanoma mutational landscape also includes genetic alterations in a number of epigenetic regulators. There is accumulating evidence that genetic alterations and changes in the expression or activity of ATP-dependent chromatin modifying enzymes, histone modifying enzymes, and readers of the histone code, can drive melanoma pathogenesis. Re-wired transcriptional programs that drive tumorigenesis often involve interactions between chromatin regulators, oncogenic signaling pathways, and transcription factors. Recent studies utilizing small molecules which inhibit chromatin remodeling enzymes and chromatin readers has provided insight into the role of epigenetic regulators in melanoma pathogenesis and shown therapeutic potential.
This Topical Collection is focused on epigenetic reprogramming by ATP-dependent chromatin remodeling enzymes, histone-modifying enzymes and readers, in the regulation of gene expression involved in melanoma metabolism, proliferation, metastasis, and the response to anti-cancer drugs. We will consider research or review manuscripts of exceptional interest on the following topics:
- Coordination of ATP-dependent chromatin remodeling enzymes, histone-modifying enzymes, and readers, transcription factors, and signaling pathways in the regulation of melanoma proliferation and metastasis.
- Genetic alterations of chromatin regulatory proteins in melanomagensis.
- Integration of chromatin modifications with melanoma metabolism.
- The use of epigenetic inhibitors as stand-alone therapeutics or in combination with targeted or immune therapies to overcome melanoma resistance.
Dr. Ivana De la Serna
Collection Editor
Manuscript Submission Information
Manuscripts should be submitted online at www.mdpi.com by registering and logging in to this website. Once you are registered, click here to go to the submission form. Manuscripts can be submitted until the deadline. All submissions that pass pre-check are peer-reviewed. Accepted papers will be published continuously in the journal (as soon as accepted) and will be listed together on the collection website. Research articles, review articles as well as short communications are invited. For planned papers, a title and short abstract (about 100 words) can be sent to the Editorial Office for announcement on this website.
Submitted manuscripts should not have been published previously, nor be under consideration for publication elsewhere (except conference proceedings papers). All manuscripts are thoroughly refereed through a single-blind peer-review process. A guide for authors and other relevant information for submission of manuscripts is available on the Instructions for Authors page. Epigenomes is an international peer-reviewed open access quarterly journal published by MDPI.
Please visit the Instructions for Authors page before submitting a manuscript.
The Article Processing Charge (APC) for publication in this open access journal is 1500 CHF (Swiss Francs).
Submitted papers should be well formatted and use good English. Authors may use MDPI's
English editing service prior to publication or during author revisions.
Keywords
- melanoma
- epigenetic
- ATP-dependent chromatin remodeling
- histone variants
- histone acetylation
- histone methylation
- bromodomains
Published Papers (3 papers)
Open AccessReview
Critical Considerations for Investigating MicroRNAs during Tumorigenesis: A Case Study in Conceptual and Contextual Nuances of miR-211-5p in Melanoma
by
Fatemeh Vand-Rajabpour, Meghan Savage, Rachel L. Belote and Robert L. Judson-Torres
Cited by 3 | Viewed by 2690
Abstract
MicroRNAs are non-coding RNAs fundamental to metazoan development and disease. Although the aberrant regulation of microRNAs during mammalian tumorigenesis is well established, investigations into the contributions of individual microRNAs are wrought with conflicting observations. The underlying cause of these inconsistencies is often attributed
[...] Read more.
MicroRNAs are non-coding RNAs fundamental to metazoan development and disease. Although the aberrant regulation of microRNAs during mammalian tumorigenesis is well established, investigations into the contributions of individual microRNAs are wrought with conflicting observations. The underlying cause of these inconsistencies is often attributed to context-specific functions of microRNAs. We propose that consideration of both context-specific factors, as well as underappreciated fundamental concepts of microRNA biology, will permit a more harmonious interpretation of ostensibly diverging data. We discuss the theory that the biological function of microRNAs is to confer robustness to specific cell states. Through this lens, we then consider the role of miR-211-5p in melanoma progression. Using literature review and meta-analyses, we demonstrate how a deep understating of domain-specific contexts is critical for moving toward a concordant understanding of miR-211-5p and other microRNAs in cancer biology.
Full article
►▼
Show Figures
Open AccessFeature PaperReview
SWI/SNF Chromatin Remodeling Enzymes in Melanoma
by
Megan R. Dreier and Ivana L. de la Serna
Cited by 10 | Viewed by 7084
Abstract
Melanoma is an aggressive malignancy that arises from the transformation of melanocytes on the skin, mucosal membranes, and uvea of the eye. SWI/SNF chromatin remodeling enzymes are multi-subunit complexes that play important roles in the development of the melanocyte lineage and in the
[...] Read more.
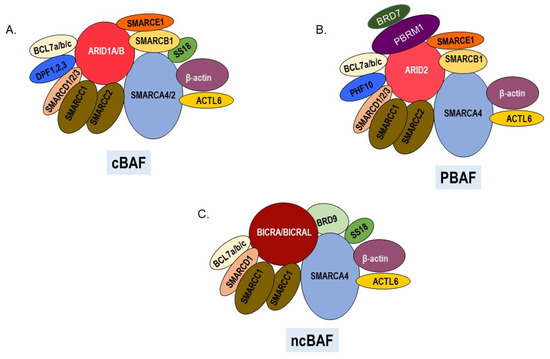
Melanoma is an aggressive malignancy that arises from the transformation of melanocytes on the skin, mucosal membranes, and uvea of the eye. SWI/SNF chromatin remodeling enzymes are multi-subunit complexes that play important roles in the development of the melanocyte lineage and in the response to ultraviolet radiation, a key environmental risk factor for developing cutaneous melanoma. Exome sequencing has revealed frequent loss of function mutations in genes encoding SWI/SNF subunits in melanoma. However, some SWI/SNF subunits have also been demonstrated to have pro-tumorigenic roles in melanoma and to affect sensitivity to therapeutics. This review summarizes studies that have implicated SWI/SNF components in melanomagenesis and have evaluated how SWI/SNF subunits modulate the response to current therapeutics.
Full article
►▼
Show Figures
Open AccessFeature PaperReview
G9a: An Emerging Epigenetic Target for Melanoma Therapy
by
Jessica L. Flesher and David E. Fisher
Cited by 8 | Viewed by 3709
Abstract
Epigenetic regulation is a crucial component of DNA maintenance and cellular identity. As our understanding of the vast array of proteins that contribute to chromatin accessibility has advanced, the role of epigenetic remodelers in disease has become more apparent. G9a is a histone
[...] Read more.
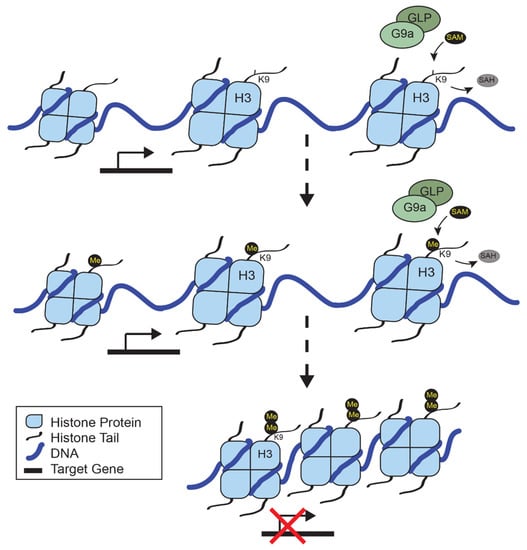
Epigenetic regulation is a crucial component of DNA maintenance and cellular identity. As our understanding of the vast array of proteins that contribute to chromatin accessibility has advanced, the role of epigenetic remodelers in disease has become more apparent. G9a is a histone methyltransferase that contributes to immune cell differentiation and function, neuronal development, and has been implicated in diseases, including cancer. In melanoma, recurrent mutations and amplifications of G9a have led to its identification as a therapeutic target. The pathways that are regulated by G9a provide an insight into relevant biomarkers for patient stratification. Future work is aided by the breadth of literature on G9a function during normal differentiation and development, along with similarities to EZH2, another histone methyltransferase that forms a synthetic lethal relationship with members of the SWI/SNF complex in certain cancers. Here, we review the literature on G9a, its role in melanoma, and lessons from EZH2 inhibitor studies.
Full article
►▼
Show Figures