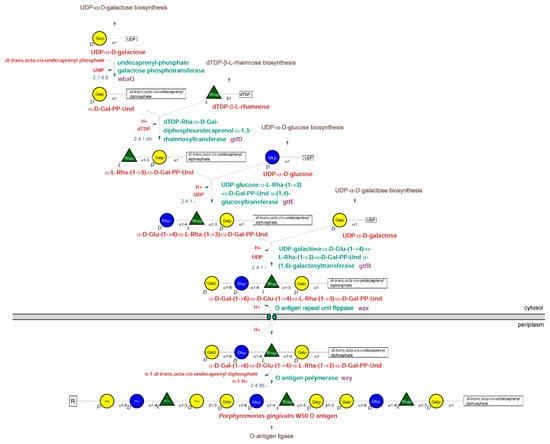
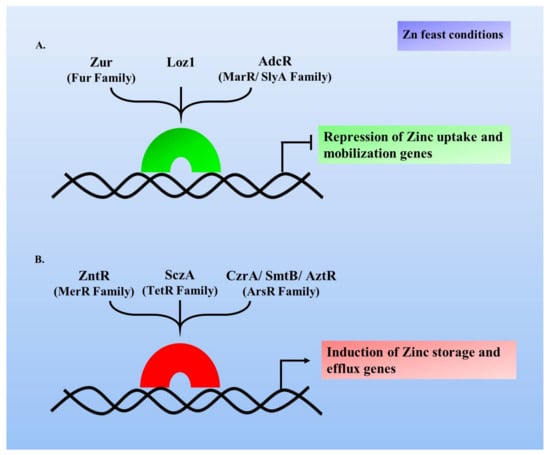
New Insights into Bacterial Pathogenesis
A topical collection in Pathogens (ISSN 2076-0817). This collection belongs to the section "Bacterial Pathogens".
Viewed by 89580Editor
Interests: host-pathogen interactions; bacterial infection; vaccines; medical mycology; innate immunity
Special Issues, Collections and Topics in MDPI journals
Topical Collection Information
Dear Colleagues,
Bacterial infections can be more or less serious and the severity of an infection depends mainly on the type of bacterium involved. Through the activation of different strategies, pathogenic bacteria are able to adhere, enter, survive, and proliferate inside host cells. If the immune defenses are not activated in time to control the infection or if an effective antibiotic treatment is not given in time, pathogenic bacteria may cross the host’s tissue barriers, gain access to internal tissues of different parts of the body, and cause severe illness.
This Topical Collection aims to provide new insights into the link between virulence, immune escape strategies, and antibiotic resistance that different microbes adopt in the context of bacterial pathogenesis. Bacterial pathogenicity is a complex and multifactorial process and the relationship between antimicrobial resistance and virulence, which often determines the ability of bacteria to cause disease, is incompletely understood. In light of this, a fundamental objective of this Topical Collection will be to gain a better understanding of the relationship between virulence and resistance that allows pathogenic bacteria to colonize and invade human tissues. This Topical Collection welcomes high-quality contributions (original research papers and reviews) that shed light on the relationship between virulence and resistance in pathogenic bacteria.
Dr. Carmelo Biondo
Collection Editor
Manuscript Submission Information
Manuscripts should be submitted online at www.mdpi.com by registering and logging in to this website. Once you are registered, click here to go to the submission form. Manuscripts can be submitted until the deadline. All submissions that pass pre-check are peer-reviewed. Accepted papers will be published continuously in the journal (as soon as accepted) and will be listed together on the collection website. Research articles, review articles as well as short communications are invited. For planned papers, a title and short abstract (about 100 words) can be sent to the Editorial Office for announcement on this website.
Submitted manuscripts should not have been published previously, nor be under consideration for publication elsewhere (except conference proceedings papers). All manuscripts are thoroughly refereed through a single-blind peer-review process. A guide for authors and other relevant information for submission of manuscripts is available on the Instructions for Authors page. Pathogens is an international peer-reviewed open access monthly journal published by MDPI.
Please visit the Instructions for Authors page before submitting a manuscript. The Article Processing Charge (APC) for publication in this open access journal is 2200 CHF (Swiss Francs). Submitted papers should be well formatted and use good English. Authors may use MDPI's English editing service prior to publication or during author revisions.
Keywords
- host–microbe interaction
- bacterial pathogenesis
- mechanisms of immunity to bacterial infection
- pathogen invasion
- virulence and resistance.