New Thiazolyl-triazole Schiff Bases: Synthesis and Evaluation of the Anti-Candida Potential
Abstract
:1. Introduction
2. Results and Discussion
2.1. Chemistry
2.2. Antifungal Activity
2.2.1. Determination of Inhibition Zone Diameters
2.2.2. Determination of Minimum Inhibitory Concentration (MIC) and Minimum Fungicidal Concentration (MFC) Values
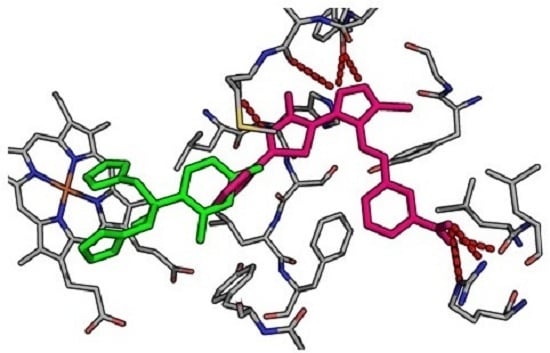
2.3. Molecular Docking
2.4. ADME and Molecular Property Prediction
3. Materials and Methods
3.1. Chemistry
3.2. Antifungal Activity Assay
3.2.1. Determination of Inhibition Zone Diameters
3.2.2. Determination of MIC and MFC Values
3.3. Molecular Docking
3.4. ADME and Molecular Property Prediction
4. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Chai, X.; Zhang, J.; Cao, Y.; Zou, Y.; Wu, Q.; Zhang, D.; Jiang, Y.; Sun, Q. Design, synthesis and molecular docking studies of novel triazole as antifungal agent. Eur. J. Med. Chem. 2011, 46, 3167–3176. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Y.; Zhang, J.; Cao, Y.; Chai, X.; Zou, Y.; Wu, Q.; Zhang, D.; Jiang, Y.; Sun, Q. Synthesis, in vitro evaluation and molecular docking studies of new triazole derivatives as antifungal agents. Bioorg. Med. Chem. Lett. 2011, 21, 4471–4475. [Google Scholar] [CrossRef] [PubMed]
- Tidwell, T.T. Hugo (Ugo) Schiff, Schiff Bases, and a Century of β-Lactam Synthesis. Angew. Chem. Int. Ed. 2008, 47, 1016–1020. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Zhao, C.P.; Ma, H.P.; Zhao, M.Y.; Xue, Y.R.; Wang, X.M. Design, synthesis and antimicrobial activities evaluation of Schiff base derived from secnidazole derivatives as potential FabH inhibitors. Bioorg. Med. Chem. 2013, 21, 3120–3126. [Google Scholar] [CrossRef] [PubMed]
- Chazin, E.L.; Sanches, P.S.; Lindgren, E.B.; Vellasco, W.T.; Pinto, L.C.; Burbano, R.M.; Yoneda, J.D.; Leal, K.Z.; Gomes, C.R.; Wardell, J.L.; et al. Synthesis and Biological Evaluation of Novel 6-Hydroxy-benzo[d][1,3]oxathiol-2-one Schiff Bases as Potential Anticancer Agents. Molecules 2015, 20, 1968–1983. [Google Scholar] [CrossRef] [PubMed]
- Barbuceanu, S.F.; Ilies, D.C.; Saramet, G.; Uivarosi, V.; Draghici, C.; Radulescu, V. Synthesis and Antioxidant Activity Evaluation of New Compounds from Hydrazinecarbothioamide and 1,2,4-Triazole Class Containing Diarylsulfone and 2,4-Difluorophenyl Moieties. Int. J. Mol. Sci. 2014, 15, 10908–10925. [Google Scholar] [CrossRef] [PubMed]
- De Souza, A.O.; Galetti, F.C.S.; Silva, C.L.; Bicalho, B.; Parma, M.M.; Fonseca, S.F.; Andrade-Neto, M. Antimycobacterial and cytotoxicity activity of synthetic and natural compounds. Quim. Nova 2007, 30, 1563–1566. [Google Scholar] [CrossRef]
- Hanif, M.; Chohan, Z.H. Design, spectral characterization and biological studies of transition metal(II) complexes with triazole Schiff bases. Spectrochim. Acta A 2013, 104, 468–476. [Google Scholar] [CrossRef] [PubMed]
- Aouad, M.R. Synthesis, Characterization and Antimicrobial Evaluation of Some New Schiff, Mannich and Acetylenic Mannich Bases Incorporating a 1,2,4-Triazole Nucleus. Molecules 2014, 19, 18897–18910. [Google Scholar] [CrossRef] [PubMed]
- Aswathanarayanappa, C.; Bheemappa, E.; Bodke, Y.D.; Krishnegowda, P.S.; Venkata, S.P.; Ningegowda, R. Synthesis and Evaluation of Antioxidant Properties of Novel 1,2,4-Triazole-Based Schiff Base Heterocycles. Arch. Pharm. 2013, 346, 922–930. [Google Scholar] [CrossRef] [PubMed]
- Anand, P.; Patil, V.M.; Sharma, V.K.; Khosa, R.L.; Masand, N. Schiff bases: A Review on Biological Insights. Int. J. Drug Des. Discov. 2012, 3, 851–868. [Google Scholar]
- Johnson, D.E.; Wolfgang, G.H. Predicting human safety: Screening and computational approaches to estimate solubility and permeability in drug discovery and development settings. Drug Discov. Today 2000, 5, 445–454. [Google Scholar] [CrossRef]
- Simiti, I.; Mureşan, A. Application of the Sommelet reaction in the series of 2-aryl-4methyl-5-chlormethyl-thiazole. Roum. Chim. 1976, 7, 1078. [Google Scholar]
- Tiperciuc, B.; Colosi, I.; Moldovan, C.; Crişan, O.; Pîrnău, A.; Vlase, L.; Duma, M.; Oniga, O. Synthesis and Evaluation of Antimicrobial Activity of Some New Hetaryl-Azoles Derivatives Obtained from 2-Aryl-4-methylthiazol-5-carbohydrazides and Isonicotinic Acid Hydrazide. J. Heterocycl. Chem. 2012, 49, 1407–1414. [Google Scholar] [CrossRef]
- Pinto, D.C.; Santos, C.M.; Silva, A.M. Advanced NMR Techniques for Structural Characterization of Heterocyclic Structures. Recent Res. Dev. Heterocycl. Chem. 2007, 37/661, 397–475. [Google Scholar]
- Alnuaimi, A.D.; O’Brien-Simpson, N.M.; Reynolds, E.C.; McCullough, M.J. Clinical isolates and laboratory reference Candida species and strains have varying abilities to form biofilms. FEMS Yeast Res. 2013, 13, 689–699. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, A.; Geschke, U. Comparative virulence of Candida albicans strains in CFW1 mice and Sprague-Dawley rats. Mycoses 1996, 39, 157–160. [Google Scholar] [CrossRef] [PubMed]
- Deorukhkar, S.C.; Saini, S.; Mathew, S. Non-albicans Candida Infection: An Emerging Threat. Interdiscip. Perspect. Infect. Dis. 2014, 2014, 615958. [Google Scholar] [PubMed]
- Movahed, E.; Yi Tan, G.M.; Munusamy, K.; Yeow, T.C.; Tay, S.T.; Wong, W.F.; Looi, C.Y. Triclosan Demonstrates Synergic Effect with Amphotericin B and Fluconazole and Induces Apoptosis-Like Cell Death in Cryptococcus neoformans. Front. Microbiol. 2016, 7. [Google Scholar] [CrossRef] [PubMed]
- Sheehan, D.J.; Hitchcock, C.A.; Sibley, C.M. Current and emerging azole antifungal agents. Clin. Microbiol. Rev. 1999, 12, 40–79. [Google Scholar] [PubMed]
- Newby, D.; Freitas, A.A.; Ghafourian, T. Decision trees to characterise the roles of permeability and solubility on the prediction of oral absorption. Eur. J. Med. Chem. 2015, 90, 751–765. [Google Scholar] [CrossRef] [PubMed]
- Azam, F.; Madi, A.M.; Ali, H.I. Molecular Docking and Prediction of Pharmacokinetic Properties of Dual Mechanism Drugs that Block MAO-B and Adenosine A2A Receptors for the Treatment of Parkinson’s Disease. J. Young Pharm. 2012, 4, 184–192. [Google Scholar] [CrossRef] [PubMed]
- Ertl, P.; Rohde, B.; Selzer, P. Fast calculation of molecular polar surface area as a sum of fragment-based contributions and its application to the prediction of drug transport properties. J. Med. Chem. 2000, 43, 3714–3717. [Google Scholar] [CrossRef] [PubMed]
- Mo, S.L.; Zhou, Z.W.; Yang, L.P.; Wei, M.Q.; Zhou, S.F. New insights into the structural features, and functional relevance of human cytochrome P4502C9. Part I. Curr. Drug Metab. 2009, 10, 1075–1126. [Google Scholar] [CrossRef] [PubMed]
- Mo, S.L.; Zhou, Z.W.; Yang, L.P.; Wei, M.Q.; Zhou, S.F. New insights into the structural features, and functional relevance of human cytochrome P4502C9. Part II. Curr. Drug Metab. 2009, 10, 1127–1150. [Google Scholar] [CrossRef] [PubMed]
- Fromm, M.F. P-glycoprotein: A defense mechanism limiting oral bioavailability and CNS accumulation of drugs. Int. J. Clin. Pharmacol. Ther. 2000, 38, 69–74. [Google Scholar] [CrossRef] [PubMed]
- Nonoyama, T.; Fukuda, R. Drug induced phospholipidosis pathological aspects and its prediction. J. Toxicol. Pathol. 2008, 21, 9–24. [Google Scholar] [CrossRef]
- Reference Method for Broth Dilution Antifungal Susceptibility Testing of Yeast; Third Informational Supplement, CLSI Document M27-S3; Clinical and Laboratory Standards Institute (CLSI): Wayne, PA, USA, 2008.
- Morris, G.M.; Huey, R.; Lindstrom, W.; Sanner, M.F.; Belew, R.K.; Goodsell, D.S.; Olson, A.J. AutoDock4 and AutoDockTools4: Automated Docking with Selective Receptor Flexibility. J. Comput. Chem. 2009, 30, 2785–2791. [Google Scholar] [CrossRef] [PubMed]
- Monk, B.C.; Tomasiak, T.M.; Keniya, M.V.; Huschmann, F.U.; Tyndall, J.D.; O’Connell, J.D.; Cannon, R.D.; McDonald, J.G.; Rodriguez, A.; Finer-Moore, J.S.; et al. Architecture of a single membrane spanning cytochrome P450 suggests constraints that orient the catalytic domain relative to a bilayer. Proc. Natl. Acad. Sci. USA 2014, 111, 3865–3870. [Google Scholar] [CrossRef] [PubMed]
- Podust, L.M.; Poulos, T.L.; Waterman, M.R. Crystal structure of cytochrome P450 14alpha-sterol demetylase (CYP51) from Mycobacterium tuberculosis in complex with azole inhibitors. Proc. Natl. Acad. Sci. USA 2001, 98, 3068–3073. [Google Scholar] [CrossRef] [PubMed]
- Lipinski, C.A.; Lombardo, F.; Dominy, B.W.; Feeney, P.J. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv. Drug Deliv. Rev. 2001, 46, 3–26. [Google Scholar] [CrossRef]
- Swiss ADME. Available online: http://www.swissadme.ch (accessed on 20 November 2016).
- Daina, A.; Zoete, V. A BOILED-Egg To Predict Gastrointestinal Absorption and Brain Penetration of Small Molecules. ChemMedChem 2016, 11, 1117–1121. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Abraham, M.H.; Lee, J.; Hersey, A.; Luscombe, N.C.; Beck, G.; Cooper, I. Rate-limited steps of human oral absorbtion and QSAR studies. Pharm. Res. 2002, 19, 1446–1457. [Google Scholar] [CrossRef] [PubMed]
- Sample Availability: Samples of the compounds B1–B15 are available from the authors.





| Cp | Inhibition Zone Diameter (mm) |
|---|---|
| Candida albicans ATCC 90028 | |
| B1 | 16 |
| B2 | 18 |
| B3 | 18 |
| B4 | 18 |
| B5 | 20 |
| B6 | 18 |
| B7 | 18 |
| B8 | 18 |
| B9 | 18 |
| B10 | 20 |
| B11 | 20 |
| B12 | 18 |
| B13 | 18 |
| B14 | 18 |
| B15 | 18 |
| Fluconazole | 25 |
| Cp | C. albicans ATCC 10231 | C. albicans ATCC 18804 | C. krusei ATCC 6258 |
|---|---|---|---|
| B1 | 62.5 | 62.5 | 62.5 |
| B2 | 62.5 | 62.5 | 62.5 |
| B3 | 62.5 | 62.5 | 62.5 |
| B4 | 62.5 | 62.5 | 62.5 |
| B5 | 62.5 | 62.5 | 31.25 |
| B6 | 62.5 | 62.5 | 62.5 |
| B7 | 62.5 | 62.5 | 62.5 |
| B8 | 62.5 | 31.25 | 62.5 |
| B9 | 62.5 | 62.5 | 62.5 |
| B10 | 15.62 | 31.25 | 31.25 |
| B11 | 62.5 | 62.5 | 62.5 |
| B12 | 62.5 | 62.5 | 62.5 |
| B13 | 62.5 | 62.5 | 62.5 |
| B14 | 62.5 | 31.25 | 62.5 |
| B15 | 62.5 | 62.5 | 62.5 |
| Fluconazole | 62.5 | 62.5 | 62.5 |
| Ketoconazole | 31.25 | 31.25 | 31.25 |
| Inoculum control | +++ | +++ | +++ |
| Broth control | No growth | No growth | No growth |
| Cp. | C. albicans ATCC 10231 | C. albicans ATCC 18804 | C. krusei ATCC 6258 |
|---|---|---|---|
| B1 | 125 | 125 | 125 |
| B2 | 125 | 125 | 125 |
| B3 | 125 | 125 | 125 |
| B4 | 125 | 125 | 125 |
| B5 | 125 | 125 | 62.5 |
| B6 | 125 | 125 | 125 |
| B7 | 125 | 125 | 125 |
| B8 | 125 | 62.5 | 125 |
| B9 | 125 | 62.5 | 125 |
| B10 | 31.25 | 62.5 | 62.5 |
| B11 | 125 | 125 | 125 |
| B12 | 125 | 125 | 125 |
| B13 | 125 | 125 | 125 |
| B14 | 125 | 62.5 | 125 |
| B15 | 125 | 125 | 62.5 |
| Fluconazole | 125 | 125 | 125 |
| Ketoconazole | 62.5 | 62.5 | 62.5 |
| Samples | Binding Affinity (kcal/mol) | Atom ID of Ligand | Interacting AA Residue | Bond Length (Å) |
|---|---|---|---|---|
| B1 | −11.39 | N1 | Met509 | 3.0 |
| N2 | Ser508 | 3.5 | ||
| N3 | Phe506 | 2.6 | ||
| B2 | −11.85 | N2 | Ser508 | 3.5 |
| N3 | Phe506 | 2.6 | ||
| B3 | −11.59 | N1 | Met509 | 2.9 |
| N2 | Ser508 | 4.4 | ||
| N3 | Phe506 | 2.6 | ||
| B4 | −11.08 | N1 | Ser382 | 3.1 |
| N2 | Ser508 | 2.9 | ||
| N3 | Ser508 | 3.2 | ||
| N3 | Phe506 | 2.5 | ||
| B5 | −11.22 | N1 | Met509 | 3.2 |
| N2 | Ser508 | 2.9 | ||
| N3 | Phe506 | 2.5 | ||
| B6 | −10.30 | N1 | Met509 | 2.9 |
| N2 | Phe506 | 2.7 | ||
| N3 | Phe506 | 2.7 | ||
| B7 | −10.92 | N1 | Met509 | 2.9 |
| N2 | Ser508 | 3.6 | ||
| N3 | Phe506 | 2.6 | ||
| B8 | −11.25 | N1 | Ser382 | 3.1 |
| N2 | Phe506 | 3.0 | ||
| N3 | Phe506 | 2.7 | ||
| Phenolic O | Arg98 | 3.1 | ||
| B9 | −10.84 | N1 | Ser382 | 3.0 |
| N2 | Ser508 | 2.9 | ||
| N3 | Ser508 | 3.2 | ||
| N3 | Phe506 | 2.6 | ||
| B10 | −12.92 | N1 | Met509 | 2.6 |
| N2 | Ser508 | 3.3 | ||
| N2 | Phe506 | 2.5 | ||
| N3 | Phe506 | 2.5 | ||
| Nitro O | Arg98 | 3.0 | ||
| Nitro O | Arg98 | 3.2 | ||
| Nitro O | Leu95 | 2.7 | ||
| B11 | −11.70 | N1 | Met509 | 2.9 |
| N2 | Ser508 | 3.5 | ||
| N3 | Phe506 | 2.6 | ||
| Nitro O | Arg98 | 3.4 | ||
| B12 | −11.17 | N1 | Ser382 | 2.8 |
| N2 | Ser508 | 2.9 | ||
| N3 | Ser508 | 3.0 | ||
| N3 | Phe506 | 2.5 | ||
| B13 | −10.93 | N1 | Ser382 | 3.3 |
| N2 | Ser508 | 3.2 | ||
| N3 | Phe506 | 2.8 | ||
| B14 | −10.45 | N1 | Ser382 | 3.1 |
| N2 | Ser508 | 2.8 | ||
| N3 | Ser508 | 3.2 | ||
| N3 | Phe506 | 2.6 | ||
| B15 | −11.18 | N1 | Met509 | 2.8 |
| N2 | Ser508 | 3.4 | ||
| N3 | Phe506 | 2.6 | ||
| Fluconazole | −7.03 | N/A | N/A | N/A |
| Comp. | tPSA a | %Abs b | MW c | RoB d | HBD e | HBA f | MR g | IlogP h (MlogP) | LogS i | CYP2D6 Inhibitor |
|---|---|---|---|---|---|---|---|---|---|---|
| Rule | ≤140 Ǻ2 | - | ≤1500 | ≤110 | ≤15 | ≤110 | 40–130 | <5 | >−4 | |
| B1 | 123 | 66.57 | 446.38 | 4 | 0 | 4 | 118.24 | 3.77 (4.12) | −8.19 | No |
| B2 | 123 | 66.57 | 446.38 | 4 | 0 | 4 | 118.24 | 3.69 (4.12) | −8.19 | No |
| B3 | 123 | 66.57 | 446.38 | 4 | 0 | 4 | 118.24 | 3.42 (4.12) | −8.19 | No |
| B4 | 123 | 66.57 | 411.93 | 4 | 0 | 4 | 113.23 | 3.86 (3.89) | −7.60 | No |
| B5 | 123 | 66.57 | 456.38 | 4 | 0 | 4 | 115.92 | 3.35 (4.01) | −7.80 | No |
| B6 | 123 | 66.57 | 395.48 | 4 | 0 | 5 | 108.18 | 3.3 (3.78) | −7.28 | No |
| B7 | 143.23 | 59.59 | 393.49 | 4 | 1 | 5 | 110.24 | 3.09 (2.85) | −6.43 | No |
| B8 | 143.23 | 59.59 | 393.49 | 4 | 1 | 5 | 110.24 | 3.12 (2.85) | −6.43 | No |
| B9 | 143.23 | 59.59 | 393.49 | 4 | 1 | 5 | 110.24 | 3.24 (2.85) | −6.43 | No |
| B10 | 168.82 | 50.76 | 422.48 | 5 | 0 | 6 | 117.04 | 2.78 (2.42) | −6.84 | No |
| B11 | 168.82 | 50.76 | 422.48 | 5 | 0 | 6 | 117.04 | 2.75 (2.42) | −6.84 | No |
| B12 | 132.23 | 63.38 | 407.51 | 5 | 0 | 5 | 114.71 | 3.98 (3.08) | −7.12 | No |
| B13 | 132.23 | 63.38 | 407.51 | 5 | 0 | 5 | 114.71 | 3.84 (3.08) | −7.12 | No |
| B14 | 151.24 | 56.82 | 383.51 | 4 | 0 | 4 | 106.09 | 3.26 (2.96) | −6.28 | No |
| B15 | 126.24 | 65.45 | 420.55 | 5 | 0 | 4 | 122.42 | 3.82 (3.3) | −7.09 | No |
© 2016 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license ( http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Stana, A.; Enache, A.; Vodnar, D.C.; Nastasă, C.; Benedec, D.; Ionuț, I.; Login, C.; Marc, G.; Oniga, O.; Tiperciuc, B. New Thiazolyl-triazole Schiff Bases: Synthesis and Evaluation of the Anti-Candida Potential. Molecules 2016, 21, 1595. https://doi.org/10.3390/molecules21111595
Stana A, Enache A, Vodnar DC, Nastasă C, Benedec D, Ionuț I, Login C, Marc G, Oniga O, Tiperciuc B. New Thiazolyl-triazole Schiff Bases: Synthesis and Evaluation of the Anti-Candida Potential. Molecules. 2016; 21(11):1595. https://doi.org/10.3390/molecules21111595
Chicago/Turabian StyleStana, Anca, Alexandra Enache, Dan Cristian Vodnar, Cristina Nastasă, Daniela Benedec, Ioana Ionuț, Cezar Login, Gabriel Marc, Ovidiu Oniga, and Brîndușa Tiperciuc. 2016. "New Thiazolyl-triazole Schiff Bases: Synthesis and Evaluation of the Anti-Candida Potential" Molecules 21, no. 11: 1595. https://doi.org/10.3390/molecules21111595
APA StyleStana, A., Enache, A., Vodnar, D. C., Nastasă, C., Benedec, D., Ionuț, I., Login, C., Marc, G., Oniga, O., & Tiperciuc, B. (2016). New Thiazolyl-triazole Schiff Bases: Synthesis and Evaluation of the Anti-Candida Potential. Molecules, 21(11), 1595. https://doi.org/10.3390/molecules21111595