Solution NMR Spectroscopy in Target-Based Drug Discovery
Abstract
:1. Introduction
2. Challenges of NMR in Drug Discovery
2.1. NMR Sample Preparation Challenges
2.1.1. Sample Preparation
2.1.2. Protein Stability
3. NMR Experiments Used for Protein-Ligand Interactions
3.1. Chemical Shift Mapping Experiments

3.1.1. Differential Chemical Shift
3.1.2. Determining the Binding Affinity

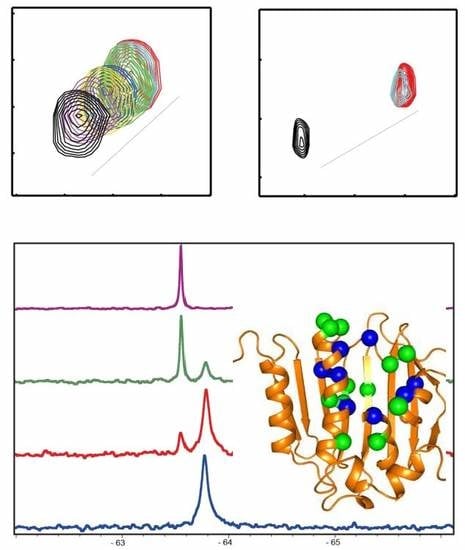
3.2. 19F-Based NMR Experiment
3.2.1. Hit Identification
3.2.2. Determining Conformational Exchanges
3.2.3. Ranking Compound Binding Affinities
4. Solution NMR in Target Engagement
5. Conclusions and Perspectives
Acknowledgments
Author Contributions
Conflicts of Interest
Abbreviations
| NMR | Nuclear magnetic resonance |
| HTS | High-throughput screening |
| FBDD | Fragment-based drug discovery |
| CSP | Chemical shift perturbation |
| HSQC | Heteronuclear single quantum coherence spectroscopy |
| E. coli | Escherichia coli |
| GST | Glutathione S-transferase |
| GyrB | gyrase B subunit |
| eParE | E. coli topoisomerase IV E subunit |
| GPCR | G protein coupled receptor |
| WNV | West Nile Virus |
References
- Bax, A. Two-dimensional NMR and protein structure. Annu. Rev. Biochem. 1989, 58, 223–256. [Google Scholar] [CrossRef] [PubMed]
- Dallmann, A.; Sattler, M. Detection of hydrogen bonds in dynamic regions of RNA by NMR spectroscopy. Curr. Protoc. Nucleic Acid Chem. 2014, 59, 1–19. [Google Scholar]
- Billeter, M.; Wagner, G.; Wuthrich, K. Solution NMR structure determination of proteins revisited. J. Biomol. NMR 2008, 42, 155–158. [Google Scholar] [CrossRef] [PubMed]
- Takeuchi, K.; Wagner, G. NMR studies of protein interactions. Curr. Opin. Struct. Biol. 2006, 16, 109–117. [Google Scholar] [CrossRef] [PubMed]
- Bax, A. Weak alignment offers new NMR opportunities to study protein structure and dynamics. Protein Sci. Publ. Protein Soc. 2003, 12, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Fesik, S.W.; Zuiderweg, E.R.; Olejniczak, E.T.; Gampe, R.T., Jr. NMR methods for determining the structures of enzyme/inhibitor complexes as an aid in drug design. Biochem. Pharmacol. 1990, 40, 161–167. [Google Scholar] [CrossRef]
- Selenko, P.; Wagner, G. Looking into live cells with in-cell NMR spectroscopy. J. Struct. Biol. 2007, 158, 244–253. [Google Scholar] [CrossRef] [PubMed]
- Tugarinov, V.; Kanelis, V.; Kay, L.E. Isotope labeling strategies for the study of high-molecular-weight proteins by solution NMR spectroscopy. Nat. Protoc. 2006, 1, 749–754. [Google Scholar] [CrossRef] [PubMed]
- Tugarinov, V.; Kay, L.E. Methyl groups as probes of structure and dynamics in NMR studies of high-molecular-weight proteins. ChemBioChem 2005, 6, 1567–1577. [Google Scholar] [CrossRef] [PubMed]
- Tugarinov, V.; Choy, W.Y.; Orekhov, V.Y.; Kay, L.E. Solution NMR-derived global fold of a monomeric 82-kDa enzyme. Proc. Natl. Acad. Sci. USA 2005, 102, 622–627. [Google Scholar] [CrossRef] [PubMed]
- Kay, L.E. NMR studies of protein structure and dynamics. J. Magn. Reson. 2005, 173, 193–207. [Google Scholar] [CrossRef] [PubMed]
- Ding, K.; Gronenborn, A.M. Sensitivity-enhanced 2D IPAP, TROSY-anti-TROSY, and E.COSY experiments: Alternatives for measuring dipolar 15N-1HN couplings. J. Magn. Reson. 2003, 163, 208–214. [Google Scholar] [CrossRef]
- Rudiger, S.; Freund, S.M.; Veprintsev, D.B.; Fersht, A.R. CRINEPT-TROSY NMR reveals p53 core domain bound in an unfolded form to the chaperone Hsp90. Proc. Natl. Acad. Sci. USA 2002, 99, 11085–11090. [Google Scholar] [CrossRef] [PubMed]
- Riek, R.; Pervushin, K.; Wuthrich, K. TROSY and CRINEPT: NMR with large molecular and supramolecular structures in solution. Trends Biochem. Sci. 2000, 25, 462–468. [Google Scholar] [CrossRef]
- Vogtherr, M.; Fiebig, K. NMR-based screening methods for lead discovery. In Modern Methods of Drug Discovery; Birkhäuser: Basel, Switzerland, 2003; pp. 183–202. [Google Scholar]
- Fejzo, J.; Lepre, C.; Xie, X. Application of NMR screening in drug discovery. Curr. Top. Med. Chem. 2003, 3, 81–97. [Google Scholar] [CrossRef] [PubMed]
- Pellecchia, M.; Sem, D.S.; Wuthrich, K. Nmr in drug discovery. Nat. Rev. Drug Discov. 2002, 1, 211–219. [Google Scholar] [CrossRef] [PubMed]
- Zega, A. NMR Methods for Identification of False Positives in Biochemical Screens. J. Med. Chem. 2017. [Google Scholar] [CrossRef] [PubMed]
- Fernandez, C.; Jahnke, W. New approaches for NMR screening in drug discovery. Drug Discov. Today Technol. 2004, 1, 277–283. [Google Scholar] [CrossRef] [PubMed]
- Hanzawa, H.; Takizawa, T. NMR screening in fragment-based drug discovery. Yakugaku Zasshi J. Pharm. Soc. Jpn. 2010, 130, 325–333. [Google Scholar] [CrossRef]
- Erlanson, D.A.; McDowell, R.S.; O’Brien, T. Fragment-Based Drug Discovery. J. Med. Chem. 2004, 47, 3463–3482. [Google Scholar] [CrossRef] [PubMed]
- Murray, C.W.; Rees, D.C. The rise of fragment-based drug discovery. Nat. Chem. 2009, 1, 187–192. [Google Scholar] [CrossRef] [PubMed]
- Baker, M. Fragment-based lead discovery grows up. Nat. Rev. Drug Discov. 2013, 12, 5–7. [Google Scholar] [CrossRef] [PubMed]
- Schade, M. NMR fragment screening: Advantages and applications. IDrugs 2006, 9, 110–113. [Google Scholar] [PubMed]
- Zartler, E.R.; Mo, H. Practical aspects of NMR-based fragment discovery. Curr. Top. Med. Chem. 2007, 7, 1592–1599. [Google Scholar] [CrossRef] [PubMed]
- Harner, M.J.; Frank, A.O.; Fesik, S.W. Fragment-based drug discovery using NMR spectroscopy. J. Biomol. NMR 2013, 56, 65–75. [Google Scholar] [CrossRef] [PubMed]
- Erlanson, D.A.; Fesik, S.W.; Hubbard, R.E.; Jahnke, W.; Jhoti, H. Twenty years on: The impact of fragments on drug discovery. Nat. Rev. Drug Discov. 2016, 15, 605–619. [Google Scholar] [CrossRef] [PubMed]
- Pelz, N.F.; Bian, Z.; Zhao, B.; Shaw, S.; Tarr, J.C.; Belmar, J.; Gregg, C.; Camper, D.V.; Goodwin, C.M.; Arnold, A.L.; et al. Discovery of 2-Indole-acylsulfonamide Myeloid Cell Leukemia 1 (Mcl-1) Inhibitors Using Fragment-Based Methods. J. Med. Chem. 2016, 59, 2054–2066. [Google Scholar] [CrossRef] [PubMed]
- Frank, A.O.; Vangamudi, B.; Feldkamp, M.D.; Souza-Fagundes, E.M.; Luzwick, J.W.; Cortez, D.; Olejniczak, E.T.; Waterson, A.G.; Rossanese, O.W.; Chazin, W.J.; et al. Discovery of a potent stapled helix peptide that binds to the 70N domain of replication protein A. J. Med. Chem. 2014, 57, 2455–2461. [Google Scholar] [CrossRef] [PubMed]
- Petros, A.M.; Dinges, J.; Augeri, D.J.; Baumeister, S.A.; Betebenner, D.A.; Bures, M.G.; Elmore, S.W.; Hajduk, P.J.; Joseph, M.K.; Landis, S.K.; et al. Discovery of a potent inhibitor of the antiapoptotic protein Bcl-xL from NMR and parallel synthesis. J. Med. Chem. 2006, 49, 656–663. [Google Scholar] [CrossRef] [PubMed]
- Campos-Olivas, R. NMR Screening and Hit Validation in Fragment Based Drug Discovery. Curr. Top. Med. Chem. 2010, 11, 43–67. [Google Scholar] [CrossRef]
- Begley, D.W.; Zheng, S.; Varani, G. Fragment-based discovery of novel thymidylate synthase leads by NMR screening and group epitope mapping. Chem. Biol. Drug Des. 2010, 76, 218–233. [Google Scholar] [CrossRef] [PubMed]
- Klages, J.; Coles, M.; Kessler, H. NMR-based screening: A powerful tool in fragment-based drug discovery. Analyst 2007, 132, 693–705. [Google Scholar] [CrossRef] [PubMed]
- Jhoti, H.; Cleasby, A.; Verdonk, M.; Williams, G. Fragment-based screening using X-ray crystallography and NMR spectroscopy. Curr. Opin. Chem. Biol. 2007, 11, 485–493. [Google Scholar] [CrossRef] [PubMed]
- Chung, F.; Tisne, C.; Lecourt, T.; Dardel, F.; Micouin, L. NMR-guided fragment-based approach for the design of tRNA(Lys3) ligands. Angew. Chem. Int. Ed. Engl. 2007, 46, 4489–4491. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huth, J.R.; Sun, C.; Sauer, D.R.; Hajduk, P.J. Utilization of NMR-derived fragment leads in drug design. Methods Enzymol. 2005, 394, 549–571. [Google Scholar] [PubMed]
- Huth, J.R.; Sun, C. Utility of NMR in lead optimization: Fragment-based approaches. Comb. Chem. High Throughput Screen. 2002, 5, 631–643. [Google Scholar] [CrossRef] [PubMed]
- Ma, R.; Wang, P.; Wu, J.; Ruan, K. Process of Fragment-Based Lead Discovery—A Perspective from NMR. Molecules 2016, 21, 854. [Google Scholar] [CrossRef] [PubMed]
- Luchinat, E.; Banci, L. In-cell NMR: A topical review. IUCrJ 2017, 4, 108–118. [Google Scholar] [CrossRef] [PubMed]
- Pastore, A.; Temussi, P.A. The Emperor’s new clothes: Myths and truths of in-cell NMR. Arch. Biochem. Biophys. 2017. [Google Scholar] [CrossRef] [PubMed]
- Aguirre, C.; Cala, O.; Krimm, I. Overview of Probing Protein-Ligand Interactions Using NMR. In Current Protocols in Protein Science; John Wiley & Sons, Inc.: Hoboken, NJ, USA, 2001. [Google Scholar]
- Goldflam, M.; Tarragó, T.; Gairí, M.; Giralt, E. NMR Studies of Protein–Ligand Interactions. In Protein NMR Techniques; Shekhtman, A., Burz, D.S., Eds.; Humana Press: Totowa, NJ, USA, 2012; pp. 233–259. [Google Scholar]
- Cala, O.; Guillière, F.; Krimm, I. NMR-based analysis of protein–ligand interactions. Anal. Bioanal. Chem. 2014, 406, 943–956. [Google Scholar] [CrossRef] [PubMed]
- Gayen, S.; Li, Q.; Kang, C.B. Solution NMR study of the transmembrane domain of single-span membrane proteins: Opportunities and strategies. Curr. Protein Pept. Sci. 2012, 13, 585–600. [Google Scholar] [CrossRef] [PubMed]
- Kang, C.; Li, Q. Solution NMR study of integral membrane proteins. Curr. Opin. Chem. Biol. 2011, 15, 560–569. [Google Scholar] [CrossRef] [PubMed]
- Wood, M.J.; Komives, E.A. Production of large quantities of isotopically labeled protein in Pichia pastoris by fermentation. J. Biomol. NMR 1999, 13, 149–159. [Google Scholar] [CrossRef] [PubMed]
- Vajpai, N.; Strauss, A.; Fendrich, G.; Cowan-Jacob, S.W.; Manley, P.W.; Grzesiek, S.; Jahnke, W. Solution Conformations and Dynamics of ABL Kinase-Inhibitor Complexes Determined by NMR Substantiate the Different Binding Modes of Imatinib/Nilotinib and Dasatinib. J. Biol. Chem. 2008, 283, 18292–18302. [Google Scholar] [CrossRef] [PubMed]
- Egorova-Zachernyuk, T.A.; Bosman, G.J.; Degrip, W.J. Uniform stable-isotope labeling in mammalian cells: Formulation of a cost-effective culture medium. Appl. Microbiol. Biotechnol. 2011, 89, 397–406. [Google Scholar] [CrossRef] [PubMed]
- Dutta, A.; Saxena, K.; Schwalbe, H.; Klein-Seetharaman, J. Isotope labeling in mammalian cells. Methods Mol. Biol. 2012, 831, 55–69. [Google Scholar] [PubMed]
- Sanders, C.R.; Sonnichsen, F. Solution NMR of membrane proteins: Practice and challenges. Magn. Reson. Chem. 2006, 44, S24–S40. [Google Scholar] [CrossRef] [PubMed]
- Takeda, M.; Kainosho, M. Cell-free protein production for NMR studies. Methods Mol. Biol. 2012, 831, 71–84. [Google Scholar] [PubMed]
- Ozawa, K.; Headlam, M.J.; Schaeffer, P.M.; Henderson, B.R.; Dixon, N.E.; Otting, G. Optimization of an Escherichia coli system for cell-free synthesis of selectively N-labelled proteins for rapid analysis by NMR spectroscopy. Eur. J. Biochem. 2004, 271, 4084–4093. [Google Scholar] [CrossRef] [PubMed]
- Apponyi, M.A.; Ozawa, K.; Dixon, N.E.; Otting, G. Cell-free protein synthesis for analysis by NMR spectroscopy. Methods Mol. Biol. 2008, 426, 257–268. [Google Scholar] [PubMed]
- Mahawaththa, M.C.; Pearce, B.J.G.; Szabo, M.; Graham, B.; Klein, C.D.; Nitsche, C.; Otting, G. Solution conformations of a linked construct of the Zika virus NS2B-NS3 protease. Antivir. Res. 2017, 142, 141–147. [Google Scholar] [CrossRef] [PubMed]
- Staunton, D.; Schlinkert, R.; Zanetti, G.; Colebrook, S.A.; Campbell, I.D. Cell-free expression and selective isotope labelling in protein NMR. Magn. Reson. Chem. 2006, 44, S2–S9. [Google Scholar] [CrossRef] [PubMed]
- Etzkorn, M.; Raschle, T.; Hagn, F.; Gelev, V.; Rice, A.J.; Walz, T.; Wagner, G. Cell-free Expressed Bacteriorhodopsin in Different Soluble Membrane Mimetics: Biophysical Properties and NMR Accessibility. Structure 2013, 21, 394–401. [Google Scholar] [CrossRef] [PubMed]
- Schwarz, D.; Junge, F.; Durst, F.; Frolich, N.; Schneider, B.; Reckel, S.; Sobhanifar, S.; Dotsch, V.; Bernhard, F. Preparative scale expression of membrane proteins in Escherichia coli-based continuous exchange cell-free systems. Nat. Protoc. 2007, 2, 2945–2957. [Google Scholar] [CrossRef] [PubMed]
- Reckel, S.; Sobhanifar, S.; Schneider, B.; Junge, F.; Schwarz, D.; Durst, F.; Lohr, F.; Guntert, P.; Bernhard, F.; Dotsch, V. Transmembrane segment enhanced labeling as a tool for the backbone assignment of alpha-helical membrane proteins. Proc. Natl. Acad. Sci. USA 2008, 105, 8262–8267. [Google Scholar] [CrossRef] [PubMed]
- Costa, S.; Almeida, A.; Castro, A.; Domingues, L. Fusion tags for protein solubility, purification and immunogenicity in Escherichia coli: The novel Fh8 system. Front. Microbiol. 2014, 5. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kay, L.E.; Ikura, M.; Tschudin, R.; Bax, A. Three-dimensional triple-resonance NMR Spectroscopy of isotopically enriched proteins. J. Magn. Reson. 2011, 213, 423–441. [Google Scholar] [CrossRef] [PubMed]
- Mobli, M.; Maciejewski, M.W.; Schuyler, A.D.; Stern, A.S.; Hoch, J.C. Sparse Sampling Methods in Multidimensional NMR. Phys. Chem. Chem. Phys. PCCP 2012, 14, 10835–10843. [Google Scholar] [CrossRef] [PubMed]
- Mobli, M.; Hoch, J.C. Nonuniform sampling and non-Fourier signal processing methods in multidimensional NMR. Prog. Nucl. Magn. Reson. Spectrosc. 2014, 83, 21–41. [Google Scholar] [CrossRef] [PubMed]
- Kazimierczuk, K.; Orekhov, V. Non-uniform sampling: Post-Fourier era of NMR data collection and processing. Magn. Reson. Chem. 2015, 53, 921–926. [Google Scholar] [CrossRef] [PubMed]
- Ying, J.; Delaglio, F.; Torchia, D.A.; Bax, A. Sparse multidimensional iterative lineshape-enhanced (SMILE) reconstruction of both non-uniformly sampled and conventional NMR data. J. Biomol. NMR 2017, 68, 101–118. [Google Scholar] [CrossRef] [PubMed]
- Sun, Z.Y.; Hyberts, S.G.; Rovnyak, D.; Park, S.; Stern, A.S.; Hoch, J.C.; Wagner, G. High-resolution aliphatic side-chain assignments in 3D HCcoNH experiments with joint H-C evolution and non-uniform sampling. J. Biomol. NMR 2005, 32, 55–60. [Google Scholar] [CrossRef] [PubMed]
- Sun, Z.Y.; Frueh, D.P.; Selenko, P.; Hoch, J.C.; Wagner, G. Fast assignment of 15N-HSQC peaks using high-resolution 3D HNcocaNH experiments with non-uniform sampling. J. Biomol. NMR 2005, 33, 43–50. [Google Scholar] [CrossRef] [PubMed]
- Hiller, S.; Garces, R.G.; Malia, T.J.; Orekhov, V.Y.; Colombini, M.; Wagner, G. Solution structure of the integral human membrane protein VDAC-1 in detergent micelles. Science 2008, 321, 1206–12010. [Google Scholar] [CrossRef] [PubMed]
- Calzolai, L.; Zahn, R. Influence of pH on NMR Structure and Stability of the Human Prion Protein Globular Domain. J. Biol. Chem. 2003, 278, 35592–35596. [Google Scholar] [CrossRef] [PubMed]
- Chagot, B.; Chazin, W.J. Solution NMR structure of Apo-calmodulin in complex with the IQ motif of human cardiac sodium channel NaV1.5. J. Mol. Biol. 2011, 406, 106–119. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Ng, H.Q.; Yoon, H.S.; Kang, C. Insight into the molecular interaction between the cyclic nucleotide-binding homology domain and the eag domain of the hERG channel. FEBS Lett. 2014, 588, 2782–2788. [Google Scholar] [CrossRef] [PubMed]
- Shin, J.; Chakraborty, G.; Bharatham, N.; Kang, C.; Tochio, N.; Koshiba, S.; Kigawa, T.; Kim, W.; Kim, K.T.; Yoon, H.S. NMR solution structure of human vaccinia-related kinase 1 (VRK1) reveals the C-terminal tail essential for its structural stability and autocatalytic activity. J. Biol. Chem. 2011, 286, 22131–22138. [Google Scholar] [CrossRef] [PubMed]
- Jaremko, L.; Jaremko, M.; Giller, K.; Becker, S.; Zweckstetter, M. Structure of the mitochondrial translocator protein in complex with a diagnostic ligand. Science 2014, 343, 1363–1366. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Wong, Y.X.; Poh, Z.Y.; Wong, Y.L.; Lee, M.Y.; Ng, H.Q.; Liu, B.; Hung, A.W.; Cherian, J.; Hill, J.; et al. NMR structural characterization of the N-terminal active domain of the gyrase B subunit from Pseudomonas aeruginosa and its complex with an inhibitor. FEBS Lett. 2015, 589, 2683–2869. [Google Scholar] [CrossRef] [PubMed]
- Leung, E.W.; Yagi, H.; Harjani, J.R.; Mulcair, M.D.; Scanlon, M.J.; Baell, J.B.; Norton, R.S. 19F NMR as a probe of ligand interactions with the iNOS binding site of SPRY domain-containing SOCS box protein 2. Chem. Biol. Drug Des. 2014, 84, 616–625. [Google Scholar] [CrossRef] [PubMed]
- Weigelt, J.; Wikstrom, M.; Schultz, J.; van Dongen, M.J. Site-selective labeling strategies for screening by NMR. Comb. Chem. High Throughput Screen. 2002, 5, 623–630. [Google Scholar] [CrossRef] [PubMed]
- Powers, R. Applications of NMR to structure-based drug design in structural genomics. J. Struct. Funct. Genom. 2002, 2, 113–123. [Google Scholar] [CrossRef]
- Hajduk, P.J.; Burns, D.J. Integration of NMR and high-throughput screening. Comb. Chem. High Throughput Screen. 2002, 5, 613–621. [Google Scholar] [CrossRef] [PubMed]
- Dalvit, C.; Flocco, M.; Veronesi, M.; Stockman, B.J. Fluorine-NMR competition binding experiments for high-throughput screening of large compound mixtures. Comb. Chem. High Throughput Screen. 2002, 5, 605–611. [Google Scholar] [CrossRef] [PubMed]
- Medek, A.; Hajduk, P.J.; Mack, J.; Fesik, S.W. The use of differential chemical shifts for determining the binding site location and orientation of protein-bound ligands. J. Am. Chem. Soc. 2000, 122, 1241–1242. [Google Scholar] [CrossRef]
- Kleckner, I.R.; Foster, M.P. An introduction to NMR-based approaches for measuring protein dynamics. Biochim. Biophys. Acta (BBA)—Proteins Proteom. 2011, 1814, 942–968. [Google Scholar] [CrossRef] [PubMed]
- Shuker, S.B.; Hajduk, P.J.; Meadows, R.P.; Fesik, S.W. Discovering high-affinity ligands for proteins: SAR by NMR. Science 1996, 274, 1531–1534. [Google Scholar] [CrossRef] [PubMed]
- Mayer, M.; Meyer, B. Characterization of Ligand Binding by Saturation Transfer Difference NMR Spectroscopy. Angew. Chem. Int. Ed. 1999, 38, 1784–1788. [Google Scholar] [CrossRef]
- Viegas, A.; Manso, J.; Nobrega, F.L.; Cabrita, E.J. Saturation-Transfer Difference (STD) NMR: A Simple and Fast Method for Ligand Screening and Characterization of Protein Binding. J. Chem. Educ. 2011, 88, 990–994. [Google Scholar] [CrossRef]
- Meyer, B.; Peters, T. NMR Spectroscopy Techniques for Screening and Identifying Ligand Binding to Protein Receptors. Angew. Chem. Int. Ed. 2003, 42, 864–890. [Google Scholar] [CrossRef] [PubMed]
- Dalvit, C.; Pevarello, P.; Tatò, M.; Veronesi, M.; Vulpetti, A.; Sundström, M. Identification of compounds with binding affinity to proteins via magnetization transfer from bulk water*. J. Biomol. NMR 2000, 18, 65–68. [Google Scholar] [CrossRef] [PubMed]
- Dalvit, C.; Fogliatto, G.; Stewart, A.; Veronesi, M.; Stockman, B. WaterLOGSY as a method for primary NMR screening: Practical aspects and range of applicability. J. Biomol. NMR 2001, 21, 349–359. [Google Scholar] [CrossRef] [PubMed]
- Balaram, P.; Bothner-By, A.A.; Dadok, J. Negative nuclear Overhuaser effects as probes of macromolecular structure. J. Am. Chem. Soc. 1972, 94, 4015–4017. [Google Scholar] [CrossRef] [PubMed]
- Balaram, P.; Bothner-By, A.A.; Breslow, E. Localization of tyrosine at the binding site of neurophysin II by negative nuclear Overhouser effects. J. Am. Chem. Soc. 1972, 94, 4017–4018. [Google Scholar] [CrossRef]
- Clore, G.M.; Gronenborn, A.M. Theory and applications of the transferred nuclear overhauser effect to the study of the conformations of small ligands bound to proteins. J. Magn. Reson. (1969) 1982, 48, 402–417. [Google Scholar] [CrossRef]
- Clore, G.M.; Gronenborn, A.M. Theory of the time dependent transferred nuclear Overhauser effect: Applications to structural analysis of ligand-protein complexes in solution. J. Magn. Reson. (1969) 1983, 53, 423–442. [Google Scholar] [CrossRef]
- Nirmala, N.R.; Lippens, G.M.; Hallenga, K. Theory and experimental results of transfer NOE experiments. II. The influence of residual mobility and relaxation centers inside the protein on the size of transfer NOEs. J. Magn. Reson. (1969) 1992, 100, 25–42. [Google Scholar] [CrossRef]
- Ni, F. Recent developments in transferred NOE methods. Prog. Nucl. Magn. Reson. Spectrosc. 1994, 26, 517–606. [Google Scholar] [CrossRef]
- Post, C.B. Exchange-transferred NOE spectroscopy and bound ligand structure determination. Curr. Opin. Struct. Biol. 2003, 13, 581–588. [Google Scholar] [CrossRef] [PubMed]
- Neuhaus, D.; Williamson, M.P. The Nuclear Overhauser Effect in Structural and Conformational Analysis, 2nd ed.; Wiley-VCH: Weinheim, Germany, 2000. [Google Scholar]
- Otting, G.; Wüthrich, K. Heteronuclear filters in two-dimensional [1H, 1H]-NMR spectroscopy: Combined use with isotope labelling for studies of macromolecular conformation and intermolecular interactions. Q. Rev. Biophys. 1990, 23, 39–96. [Google Scholar] [CrossRef] [PubMed]
- Ikura, M.; Bax, A. Isotope-filtered 2D NMR of a protein-peptide complex: Study of a skeletal muscle myosin light chain kinase fragment bound to calmodulin. J. Am. Chem. Soc. 1992, 114, 2433–2440. [Google Scholar] [CrossRef]
- Zwahlen, C.; Legault, P.; Vincent, S.J.F.; Greenblatt, J.; Konrat, R.; Kay, L.E. Methods for Measurement of Intermolecular NOEs by Multinuclear NMR Spectroscopy: Application to a Bacteriophage λ N-Peptide/boxB RNA Complex. J. Am. Chem. Soc. 1997, 119, 6711–6721. [Google Scholar] [CrossRef]
- Breeze, A.L. Isotope-filtered NMR methods for the study of biomolecular structure and interactions. Prog. Nucl. Magn. Reson. Spectrosc. 2000, 36, 323–372. [Google Scholar] [CrossRef]
- Vakonakis, I.; Sun, J.; Wu, T.; Holzenburg, A.; Golden, S.S.; LiWang, A.C. NMR structure of the KaiC-interacting C-terminal domain of KaiA, a circadian clock protein: Implications for KaiA–KaiC interaction. Proc. Natl. Acad. Sci. USA 2004, 101, 1479–1484. [Google Scholar] [CrossRef] [PubMed]
- Koenig, B.W.; Mitchell, D.C.; Konig, S.; Grzesiek, S.; Litman, B.J.; Bax, A. Measurement of dipolar couplings in a transducin peptide fragment weakly bound to oriented photo-activated rhodopsin. J. Biomol. NMR 2000, 16, 121–125. [Google Scholar] [CrossRef] [PubMed]
- Bolon, P.J.; Al-Hashimi, H.M.; Prestegard, J.H. Residual dipolar coupling derived orientational constraints on ligand geometry in a 53 kDa protein-ligand complex11Edited by P.E. Wright. J. Mol. Biol. 1999, 293, 107–115. [Google Scholar] [CrossRef] [PubMed]
- Lipsitz, R.S.; Tjandra, N. Residual Dipolar Couplings in NMR Structure Analysis. Annu. Rev. Biophys. Biomol. Struct. 2004, 33, 387–413. [Google Scholar] [CrossRef] [PubMed]
- Dalvit, C.; Fagerness, P.E.; Hadden, D.T.A.; Sarver, R.W.; Stockman, B.J. Fluorine-NMR Experiments for High-Throughput Screening: Theoretical Aspects, Practical Considerations, and Range of Applicability. J. Am. Chem. Soc. 2003, 125, 7696–7703. [Google Scholar] [CrossRef] [PubMed]
- Dalvit, C. Ligand- and substrate-based 19F NMR screening: Principles and applications to drug discovery. Prog. Nucl. Magn. Reson. Spectrosc. 2007, 51, 243–271. [Google Scholar] [CrossRef]
- Horst, R.; Liu, J.J.; Stevens, R.C.; Wuthrich, K. beta(2)-adrenergic receptor activation by agonists studied with (1)(9)F NMR spectroscopy. Angew. Chem. Int. Ed. Engl. 2013, 52, 10762–10765. [Google Scholar] [CrossRef] [PubMed]
- Gee, C.T.; Arntson, K.E.; Urick, A.K.; Mishra, N.K.; Hawk, L.M.L.; Wisniewski, A.J.; Pomerantz, W.C.K. Protein-observed 19F-NMR for fragment screening, affinity quantification and druggability assessment. Nat. Protoc. 2016, 11, 1414–1427. [Google Scholar] [CrossRef] [PubMed]
- Hoang, J.; Prosser, R.S. Conformational selection and functional dynamics of calmodulin: A (19)F nuclear magnetic resonance study. Biochemistry 2014, 53, 5727–5736. [Google Scholar] [CrossRef] [PubMed]
- Nakanishi, T.; Miyazawa, M.; Sakakura, M.; Terasawa, H.; Takahashi, H.; Shimada, I. Determination of the interface of a large protein complex by transferred cross-saturation measurements. J. Mol. Biol. 2002, 318, 245–249. [Google Scholar] [CrossRef]
- Ueda, T.; Takeuchi, K.; Nishida, N.; Stampoulis, P.; Kofuku, Y.; Osawa, M.; Shimada, I. Cross-saturation and transferred cross-saturation experiments. Q. Rev. Biophys. 2014, 47, 143–187. [Google Scholar] [CrossRef] [PubMed]
- Lee, M.D.; Loh, C.T.; Shin, J.; Chhabra, S.; Dennis, M.L.; Otting, G.; Swarbrick, J.D.; Graham, B. Compact, hydrophilic, lanthanide-binding tags for paramagnetic NMR spectroscopy. Chem. Sci. 2015, 6, 2614–2624. [Google Scholar] [CrossRef]
- Chen, W.N.; Loscha, K.V.; Nitsche, C.; Graham, B.; Otting, G. The dengue virus NS2B-NS3 protease retains the closed conformation in the complex with BPTI. FEBS Lett. 2014, 588, 2206–2211. [Google Scholar] [CrossRef] [PubMed]
- Paterson, Y.; Englander, S.; Roder, H. An antibody binding site on cytochrome c defined by hydrogen exchange and two-dimensional NMR. Science 1990, 249, 755–759. [Google Scholar] [CrossRef] [PubMed]
- Wildes, D.; Marqusee, S. Hydrogen exchange and ligand binding: Ligand-dependent and ligand-independent protection in the Src SH3 domain. Protein Sci. 2005, 14, 81–88. [Google Scholar] [CrossRef] [PubMed]
- Jahnke, W.; Floersheim, P.; Ostermeier, C.; Zhang, X.; Hemmig, R.; Hurth, K.; Uzunov, D.P. NMR Reporter Screening for the Detection of High-Affinity Ligands. Angew. Chem. Int. Ed. 2002, 41, 3420–3423. [Google Scholar] [CrossRef]
- Shortridge, M.D.; Hage, D.S.; Harbison, G.S.; Powers, R. Estimating protein-ligand binding affinity using high-throughput screening by NMR. J. Comb. Chem. 2008, 10, 948–958. [Google Scholar] [PubMed]
- Mulder, F.A.A.; Skrynnikov, N.R.; Hon, B.; Dahlquist, F.W.; Kay, L.E. Measurement of Slow (μs-ms) Time Scale Dynamics in Protein Side Chains by 15N Relaxation Dispersion NMR Spectroscopy: Application to Asn and Gln Residues in a Cavity Mutant of T4 Lysozyme. J. Am. Chem. Soc. 2001, 123, 967–975. [Google Scholar] [CrossRef] [PubMed]
- Ferreon, J.C.; Hilser, V.J. Ligand-induced changes in dynamics in the RT loop of the C-terminal SH3 domain of Sem-5 indicate cooperative conformational coupling. Protein Sci. 2003, 12, 982–996. [Google Scholar] [PubMed]
- Price, W.S.; Elwinger, F.; Vigouroux, C.; Stilbs, P. PGSE-WATERGATE, a new tool for NMR diffusion-based studies of ligand–macromolecule binding. Magn. Reson. Chem. 2002, 40, 391–395. [Google Scholar]
- Williamson, M.P. Using chemical shift perturbation to characterise ligand binding. Prog. Nucl. Magn. Reson. Spectrosc. 2013, 73, 1–16. [Google Scholar] [PubMed]
- Zhang, O.; Forman-Kay, J.D.; Shortle, D.; Kay, L.E. Triple-resonance NOESY-based experiments with improved spectral resolution: Applications to structural characterization of unfolded, partially folded and folded proteins. J. Biomol. NMR 1997, 9, 181–200. [Google Scholar] [PubMed]
- Tugarinov, V.; Kay, L.E. Ile, Leu, and Val methyl assignments of the 723-residue malate synthase G using a new labeling strategy and novel NMR methods. J. Am. Chem. Soc. 2003, 125, 13868–13878. [Google Scholar] [PubMed]
- Tugarinov, V.; Hwang, P.M.; Kay, L.E. Nuclear magnetic resonance spectroscopy of high-molecular-weight proteins. Annu. Rev. Biochem. 2004, 73, 107–146. [Google Scholar] [CrossRef] [PubMed]
- O’Connell, M.R.; Gamsjaeger, R.; Mackay, J.P. The structural analysis of protein-protein interactions by NMR spectroscopy. Proteomics 2009, 9, 5224–5232. [Google Scholar] [CrossRef] [PubMed]
- Gardner, K.H.; Kay, L.E. The use of 2H, 13C, 15N multidimensional NMR to study the structure and dynamics of proteins. Annu. Rev. Biophys. Biomol. Struct. 1998, 27, 357–406. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.M.; Gayen, S.; Kang, C.; Joy, J.; Huang, Q.; Chen, A.S.; Wee, J.L.; Ang, M.J.; Lim, H.A.; Hung, A.W.; et al. NMR Analysis of a Novel Enzymatically Active Unlinked Dengue NS2B-NS3 Protease Complex. J. Biol. Chem. 2013, 288, 12891–12900. [Google Scholar] [CrossRef] [PubMed]
- Salzmann, M.; Pervushin, K.; Wider, G.; Senn, H.; Wuthrich, K. TROSY in triple-resonance experiments: New perspectives for sequential NMR assignment of large proteins. Proc. Natl. Acad. Sci. USA 1998, 95, 13585–13590. [Google Scholar] [CrossRef] [PubMed]
- Phoo, W.W.; Li, Y.; Zhang, Z.; Lee, M.Y.; Loh, Y.R.; Tan, Y.B.; Ng, E.Y.; Lescar, J.; Kang, C.; Luo, D. Structure of the NS2B-NS3 protease from Zika virus after self-cleavage. Nat. Commun. 2016, 7, 13410. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Wong, Y.L.; Lee, M.Y.; Li, Q.; Wang, Q.Y.; Lescar, J.; Shi, P.Y.; Kang, C. Secondary Structure and Membrane Topology of the Full-Length Dengue Virus NS4B in Micelles. Angew. Chem. Int. Ed. Engl. 2016, 55, 12068–12072. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Wong, Y.L.; Lee, M.Y.; Li, Y.; Kang, C. Solution structure of the transmembrane domain of the mouse erythropoietin receptor in detergent micelles. Sci. Rep. 2015, 5, 13586. [Google Scholar] [CrossRef] [PubMed]
- Weigelt, J. Single scan, sensitivity- and gradient-enhanced TROSY for multidimensional NMR experiments. J. Am. Chem. Soc. 1998, 120, 10778–10779. [Google Scholar] [CrossRef]
- Li, Y.; Ng, H.Q.; Ngo, A.; Liu, S.; Tan, Y.W.; Kwek, P.Z.; Hung, A.W.; Joy, J.; Hill, J.; Keller, T.H.; et al. Backbone resonance assignments for the SET domain of human methyltransferase NSD3 in complex with its cofactor. Biomol. NMR Assign. 2017. [Google Scholar] [CrossRef] [PubMed]
- Muchmore, S.W.; Sattler, M.; Liang, H.; Meadows, R.P.; Harlan, J.E.; Yoon, H.S.; Nettesheim, D.; Chang, B.S.; Thompson, C.B.; Wong, S.L.; et al. X-ray and NMR structure of human Bcl-xL, an inhibitor of programmed cell death. Nature 1996, 381, 335–341. [Google Scholar] [CrossRef] [PubMed]
- De la Cruz, L.; Nguyen, T.H.; Ozawa, K.; Shin, J.; Graham, B.; Huber, T.; Otting, G. Binding of low molecular weight inhibitors promotes large conformational changes in the dengue virus NS2B-NS3 protease: Fold analysis by pseudocontact shifts. J. Am. Chem. Soc. 2011, 133, 19205–19215. [Google Scholar] [CrossRef] [PubMed]
- Su, X.C.; Ozawa, K.; Qi, R.; Vasudevan, S.G.; Lim, S.P.; Otting, G. NMR analysis of the dynamic exchange of the NS2B cofactor between open and closed conformations of the West Nile virus NS2B-NS3 protease. PLoS Negl. Trop. Dis. 2009, 3, e561. [Google Scholar] [CrossRef] [PubMed]
- Ekonomiuk, D.; Su, X.C.; Ozawa, K.; Bodenreider, C.; Lim, S.P.; Yin, Z.; Keller, T.H.; Beer, D.; Patel, V.; Otting, G.; et al. Discovery of a non-peptidic inhibitor of west nile virus NS3 protease by high-throughput docking. PLoS Negl. Trop. Dis. 2009, 3, e356. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kang, C.; Gayen, S.; Wang, W.; Severin, R.; Chen, A.S.; Lim, H.A.; Chia, C.S.; Schuller, A.; Doan, D.N.; Poulsen, A.; et al. Exploring the binding of peptidic West Nile virus NS2B-NS3 protease inhibitors by NMR. Antivir. Res. 2013, 97, 137–144. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Zhang, Z.; Phoo, W.W.; Loh, Y.R.; Wang, W.; Liu, S.; Chen, M.W.; Hung, A.W.; Keller, T.H.; Luo, D.; et al. Structural Dynamics of Zika Virus NS2B-NS3 Protease Binding to Dipeptide Inhibitors. Structure 2017, 25, 1242–1250.e3. [Google Scholar] [CrossRef] [PubMed]
- Waudby, C.A.; Ramos, A.; Cabrita, L.D.; Christodoulou, J. Two-Dimensional NMR Lineshape Analysis. Sci. Rep. 2016, 6, 24826. [Google Scholar] [CrossRef] [PubMed]
- Kang, C.; Bharatham, N.; Chia, J.; Mu, Y.; Baek, K.; Yoon, H.S. The natively disordered loop of Bcl-2 undergoes phosphorylation-dependent conformational change and interacts with Pin1. PLoS ONE 2012, 7, e52047. [Google Scholar] [CrossRef] [PubMed]
- Arai, M.; Ferreon, J.C.; Wright, P.E. Quantitative Analysis of Multisite Protein-Ligand Interactions by NMR: Binding of Intrinsically Disordered p53 Transactivation Subdomains with the TAZ2 Domain of CBP. J. Am. Chem. Soc. 2012, 134, 3792–3803. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Li, Y.; Loh, Y.R.; Phoo, W.W.; Hung, A.W.; Kang, C.; Luo, D. Crystal structure of unlinked NS2B-NS3 protease from Zika virus. Science 2016, 354, 1597–1600. [Google Scholar] [CrossRef] [PubMed]
- D’Silva, L.; Ozdowy, P.; Krajewski, M.; Rothweiler, U.; Singh, M.; Holak, T.A. Monitoring the Effects of Antagonists on Protein—Protein Interactions with NMR Spectroscopy. J. Am. Chem. Soc. 2005, 127, 13220–13226. [Google Scholar] [CrossRef] [PubMed]
- Śledź, P.; Abell, C.; Ciulli, A. Ligand-Observed NMR in Fragment-Based Approaches. In NMR of Biomolecules; Wiley-VCH Verlag GmbH & Co. KGaA: Weinheim, Germany, 2012; pp. 264–280. [Google Scholar]
- Mashalidis, E.H.; Śledź, P.; Lang, S.; Abell, C. A three-stage biophysical screening cascade for fragment-based drug discovery. Nat. Protoc. 2013, 8, 2309–2324. [Google Scholar] [CrossRef] [PubMed]
- Pellecchia, M.; Bertini, I.; Cowburn, D.; Dalvit, C.; Giralt, E.; Jahnke, W.; James, T.L.; Homans, S.W.; Kessler, H.; Luchinat, C.; et al. Perspectives on NMR in drug discovery: A technique comes of age. Nat. Rev. Drug Discov. 2008, 7, 738–745. [Google Scholar] [CrossRef] [PubMed]
- Schaumburg, K.; Deverell, C. Fluorine-19 nuclear magnetic resonance chemical shift of hydrofluoric acid in normal water and heavy water solutions. J. Am. Chem. Soc. 1968, 90, 2495–2499. [Google Scholar] [CrossRef]
- Gerig, J.T. Fluorine NMR of proteins. Prog. Nucl. Magn. Reson. Spectrosc. 1994, 26, 293–370. [Google Scholar] [CrossRef]
- Didenko, T.; Liu, J.J.; Horst, R.; Stevens, R.C.; Wuthrich, K. Fluorine-19 NMR of integral membrane proteins illustrated with studies of GPCRs. Curr. Opin. Struct. Biol. 2013, 23, 740–747. [Google Scholar] [CrossRef] [PubMed]
- Cobb, S.L.; Murphy, C.D. 19F NMR applications in chemical biology. J. Fluor. Chem. 2009, 130, 132–143. [Google Scholar] [CrossRef]
- Kitevski-LeBlanc, J.L.; Prosser, R.S. Current applications of 19F NMR to studies of protein structure and dynamics. Prog. Nucl. Magn. Reson. Spectrosc. 2012, 62, 1–33. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.J.; Horst, R.; Katritch, V.; Stevens, R.C.; Wuthrich, K. Biased signaling pathways in beta2-adrenergic receptor characterized by 19F-NMR. Science 2012, 335, 1106–1110. [Google Scholar] [CrossRef] [PubMed]
- Jackson, J.C.; Hammill, J.T.; Mehl, R.A. Site-Specific Incorporation of a 19F-Amino Acid into Proteins as an NMR Probe for Characterizing Protein Structure and Reactivity. J. Am. Chem. Soc. 2007, 129, 1160–1166. [Google Scholar] [CrossRef] [PubMed]
- Hammill, J.T.; Miyake-Stoner, S.; Hazen, J.L.; Jackson, J.C.; Mehl, R.A. Preparation of site-specifically labeled fluorinated proteins for 19F-NMR structural characterization. Nat. Protoc. 2007, 2, 2601–2607. [Google Scholar] [CrossRef] [PubMed]
- O’Hagan, D.; Schaffrath, C.; Cobb, S.L.; Hamilton, J.T.; Murphy, C.D. Biochemistry: Biosynthesis of an organofluorine molecule. Nature 2002, 416, 279. [Google Scholar] [CrossRef] [PubMed]
- Leone, M.; Rodriguez-Mias, R.A.; Pellecchia, M. Selective incorporation of 19F-labeled Trp side chains for NMR-spectroscopy-based ligand-protein interaction studies. ChemBioChem 2003, 4, 649–650. [Google Scholar] [CrossRef] [PubMed]
- Norton, R.; Leung, E.; Chandrashekaran, I.; MacRaild, C. Applications of 19F-NMR in Fragment-Based Drug Discovery. Molecules 2016, 21, 860. [Google Scholar] [CrossRef] [PubMed]
- Ye, L.; Van Eps, N.; Zimmer, M.; Ernst, O.P.; Scott Prosser, R. Activation of the A2A adenosine G-protein-coupled receptor by conformational selection. Nature 2016, 533, 265–268. [Google Scholar] [CrossRef] [PubMed]
- Zhu, L.; Yang, J.; Li, H.; Sun, H.; Liu, J.; Wang, J. Conformational change study of dengue virus NS2B-NS3 protease using 19F NMR spectroscopy. Biochem. Biophys. Res. Commun. 2015, 461, 677–680. [Google Scholar] [CrossRef] [PubMed]
- Dalvit, C.; Flocco, M.; Knapp, S.; Mostardini, M.; Perego, R.; Stockman, B.J.; Veronesi, M.; Varasi, M. High-throughput NMR-based screening with competition binding experiments. J. Am. Chem. Soc. 2002, 124, 7702–7709. [Google Scholar] [CrossRef] [PubMed]
- Siriwardena, A.H.; Tian, F.; Noble, S.; Prestegard, J.H. A straightforward NMR-spectroscopy-based method for rapid library screening. Angew. Chem. Int. Ed. Engl. 2002, 41, 3454–3457. [Google Scholar] [CrossRef]
- Li, Y.; Wong, Y.L.; Ng, F.M.; Liu, B.; Wong, Y.X.; Poh, Z.Y.; Then, S.W.; Lee, M.Y.; Ng, H.Q.; Hung, A.W.; et al. Characterization of the interaction between Escherichia coli topoisomerase IV E subunit and an ATP competitive inhibitor. Biochem. Biophys. Res. Commun. 2015, 467, 961–966. [Google Scholar] [CrossRef] [PubMed]
- Kang, C.; Li, Y.; Cherian, J.; Liu, B.; Ng, H.Q.; Lee, M.Y.; Binte Ahmad, N.H.; Poh, Z.Y.; Wong, Y.X.; Huang, Q.; et al. Biophysical Studies of Bacterial Topoisomerases Substantiate Their Binding Modes to an Inhibitor. Biophys. J. 2015, 109, 1969–1977. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Wong, Y.L.; Ng, F.M.; Liu, B.; Wong, Y.X.; Poh, Z.Y.; Liu, S.; Then, S.W.; Lee, M.Y.; Ng, H.Q.; et al. Escherichia coli topoisomerase IV E subunit and an inhibitor binding mode revealed by NMR spectroscopy. J Biol. Chem. 2016, 291, 17743–17753. [Google Scholar] [CrossRef] [PubMed]
- Chen, G.-Y.; Ng, F.M.; Tan, Y.W.; Poulsen, A.; Seetoh, W.; Lin, G.; Kang, C.; Then, S.W.; Ahmad, N.H.; Wong, Y.L.; et al. Application of Fragment-Based Drug Discovery against DNA Gyrase B. ChemPlusChem 2015, 80, 1250–1254. [Google Scholar] [CrossRef]
- Durham, T.B.; Blanco, M.-J. Target Engagement in Lead Generation. Bioorgan. Med. Chem. Lett. 2015, 25, 998–1008. [Google Scholar] [CrossRef] [PubMed]
- Molina, D.M.; Jafari, R.; Ignatushchenko, M.; Seki, T.; Larsson, E.A.; Dan, C.; Sreekumar, L.; Cao, Y.; Nordlund, P. Monitoring drug target engagement in cells and tissues using the cellular thermal shift assay. Science 2013, 341, 84–87. [Google Scholar] [CrossRef] [PubMed]
- Jafari, R.; Almqvist, H.; Axelsson, H.; Ignatushchenko, M.; Lundback, T.; Nordlund, P.; Martinez Molina, D. The cellular thermal shift assay for evaluating drug target interactions in cells. Nat. Protoc. 2014, 9, 2100–2122. [Google Scholar] [CrossRef] [PubMed]
- Dubach, J.M.; Kim, E.; Yang, K.; Cuccarese, M.; Giedt, R.J.; Meimetis, L.G.; Vinegoni, C.; Weissleder, R. Quantitating drug-target engagement in single cells in vitro and in vivo. Nat. Chem. Biol. 2017, 13, 168–173. [Google Scholar] [CrossRef] [PubMed]
- Reckel, S.; Lohr, F.; Dotsch, V. In-cell NMR spectroscopy. Chembiochem. Eur. J. Chem. Biol. 2005, 6, 1601–1606. [Google Scholar] [CrossRef] [PubMed]
- Serber, Z.; Corsini, L.; Durst, F.; Dotsch, V. In-cell NMR spectroscopy. Methods Enzymol. 2005, 394, 17–41. [Google Scholar] [PubMed]
- Sakakibara, D.; Sasaki, A.; Ikeya, T.; Hamatsu, J.; Hanashima, T.; Mishima, M.; Yoshimasu, M.; Hayashi, N.; Mikawa, T.; Walchli, M.; et al. Protein structure determination in living cells by in-cell NMR spectroscopy. Nature 2009, 458, 102–105. [Google Scholar] [CrossRef] [PubMed]
- Ikeya, T.; Sasaki, A.; Sakakibara, D.; Shigemitsu, Y.; Hamatsu, J.; Hanashima, T.; Mishima, M.; Yoshimasu, M.; Hayashi, N.; Mikawa, T.; et al. NMR protein structure determination in living E. coli cells using nonlinear sampling. Nat. Protoc. 2010, 5, 1051–1060. [Google Scholar] [CrossRef] [PubMed]
- Burz, D.S.; Dutta, K.; Cowburn, D.; Shekhtman, A. Mapping structural interactions using in-cell NMR spectroscopy (STINT-NMR). Nat. Methods 2006, 3, 91–93. [Google Scholar] [CrossRef] [PubMed]
- Dedmon, M.M.; Patel, C.N.; Young, G.B.; Pielak, G.J. FlgM gains structure in living cells. Proc. Natl. Acad. Sci. USA 2002, 99, 12681–12684. [Google Scholar] [CrossRef] [PubMed]
- McNulty, B.C.; Young, G.B.; Pielak, G.J. Macromolecular crowding in the Escherichia coli periplasm maintains alpha-synuclein disorder. J. Mol. Biol. 2006, 355, 893–897. [Google Scholar] [CrossRef] [PubMed]
- Sakai, T.; Tochio, H.; Tenno, T.; Ito, Y.; Kokubo, T.; Hiroaki, H.; Shirakawa, M. In-cell NMR spectroscopy of proteins inside Xenopus laevis oocytes. J. Biomol. NMR 2006, 36, 179–188. [Google Scholar] [CrossRef] [PubMed]
- Thongwichian, R.; Selenko, P. In-cell NMR in Xenopus laevis oocytes. Methods Mol. Biol. 2012, 895, 33–41. [Google Scholar] [PubMed]
- Inomata, K.; Ohno, A.; Tochio, H.; Isogai, S.; Tenno, T.; Nakase, I.; Takeuchi, T.; Futaki, S.; Ito, Y.; Hiroaki, H.; et al. High-resolution multi-dimensional NMR spectroscopy of proteins in human cells. Nature 2009, 458, 106–109. [Google Scholar] [CrossRef] [PubMed]
- Mercatelli, E.; Barbieri, L.; Luchinat, E.; Banci, L. Direct structural evidence of protein redox regulation obtained by in-cell NMR. Biochim. Biophys. Acta 2016, 1863, 198–204. [Google Scholar] [CrossRef] [PubMed]
- Barbieri, L.; Luchinat, E.; Banci, L. Protein interaction patterns in different cellular environments are revealed by in-cell NMR. Sci. Rep. 2015, 5, 14456. [Google Scholar] [CrossRef] [PubMed]
- Luchinat, E.; Barbieri, L.; Rubino, J.T.; Kozyreva, T.; Cantini, F.; Banci, L. In-cell NMR reveals potential precursor of toxic species from SOD1 fALS mutants. Nat. Commun. 2014, 5, 5502. [Google Scholar] [CrossRef] [PubMed]
- Barbieri, L.; Luchinat, E.; Banci, L. Characterization of proteins by in-cell NMR spectroscopy in cultured mammalian cells. Nat. Protoc. 2016, 11, 1101–1111. [Google Scholar] [CrossRef] [PubMed]
- Selenko, P.; Frueh, D.P.; Elsaesser, S.J.; Haas, W.; Gygi, S.P.; Wagner, G. In situ observation of protein phosphorylation by high-resolution NMR spectroscopy. Nat. Struct. Mol. Biol. 2008, 15, 321–329. [Google Scholar] [CrossRef] [PubMed]
- Xie, J.; Thapa, R.; Reverdatto, S.; Burz, D.S.; Shekhtman, A. Screening of Small Molecule Interactor Library by Using In-Cell NMR Spectroscopy (SMILI-NMR). J. Med. Chem. 2009, 52, 3516–3522. [Google Scholar] [CrossRef] [PubMed]
- Rahman, S.; Byun, Y.; Hassan, M.I.; Kim, J.; Kumar, V. Towards understanding cellular structure biology: In-cell NMR. Biochim. Biophys. Acta 2017, 1865, 547–557. [Google Scholar] [CrossRef] [PubMed]




| Experiments | Protein Labeling | References |
|---|---|---|
| 1H-15N/13C-HSQC | 15N/13C | [17,81] |
| Saturation Transfer Difference | NA 1 | [82,83,84] |
| WaterLOGSY | NA | [85,86] |
| Transferred NOESY Experiment | 13C, 15N, or NA | [87,88,89,90,91,92,93,94] |
| Filtered NOESY | 13C, 15N | [95,96,97,98,99] |
| Residue Dipolar Coupling | 15N, 13C, or NA | [100,101,102] |
| Ligand-observed 19F-NMR | NA | [103,104] |
| Protein-observed 19F-NMR | 19F | [105,106,107] |
| Cross-saturation | 15N | [108,109] |
| Paramagnetic Relaxation Enhancement | 15N, 13C/15N | [50,110,111] |
| H-D exchange | 15N | [112,113] |
| NMR reporter screening/competition assay | NA | [114,115] |
| Relaxation and relaxation dispersion | 15N | [116,117,118] |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, Y.; Kang, C. Solution NMR Spectroscopy in Target-Based Drug Discovery. Molecules 2017, 22, 1399. https://doi.org/10.3390/molecules22091399
Li Y, Kang C. Solution NMR Spectroscopy in Target-Based Drug Discovery. Molecules. 2017; 22(9):1399. https://doi.org/10.3390/molecules22091399
Chicago/Turabian StyleLi, Yan, and Congbao Kang. 2017. "Solution NMR Spectroscopy in Target-Based Drug Discovery" Molecules 22, no. 9: 1399. https://doi.org/10.3390/molecules22091399
APA StyleLi, Y., & Kang, C. (2017). Solution NMR Spectroscopy in Target-Based Drug Discovery. Molecules, 22(9), 1399. https://doi.org/10.3390/molecules22091399