Chemical Reporters and Their Bioorthogonal Reactions for Labeling Protein O-GlcNAcylation
Abstract
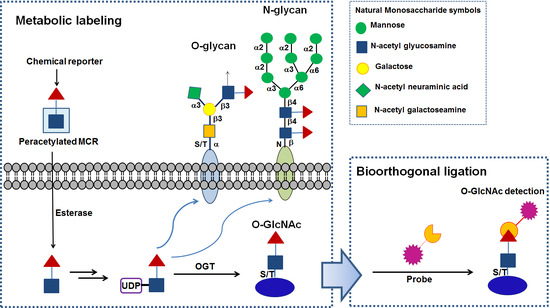
:1. Introduction
2. Metabolic Chemical Reporters and Their Chemistries for Labeling O-GlcNAc Proteins
2.1. Azide- or Terminal Alkyne-Containing Metabolic Chemical Reporters
2.2. Chemoselective Reactions Involving Azide-Functionality
2.3. Cycloalkene-Containing Metabolic Chemical Reporter
2.4. Chemoselective Ligation Reactions Involving Strained Alkene (or Alkyne) with Tetrazine through iEDDA Reaction
2.5. Diazirine-Containing Metabolic Chemical Reporter
2.6. Chemoselective Reactions Involving Diazirine-Functionality
2.7. Important Aspects of Consideration for Metabolic Chemical Reporters
3. Chemical Reporters and Their Chemistries for Labeling O-GlcNAc In Vitro
3.1. Ketone-Functionalized Chemical Reporter for Labeling O-GlcNAc In Vitro
3.2. Chemoselective Reactions Involving Ketone-Functionality
3.3. Azide-Functionalized Chemical Reporter for Labeling O-GlcNAc In Vitro
4. Conclusions and Future Perspective
Funding
Conflicts of Interest
References
- Holt, G.D.; Hart, G.W. The subcellular distribution of terminal N-acetylglucosamine moieties. Localization of a novel protein-saccharide linkage, O-linked GlcNAc. J. Biol. Chem. 1986, 261, 8049–8057. [Google Scholar] [PubMed]
- Wang, X.; Yuan, Z.-F.; Fan, J.; Karch, K.R.; Ball, L.E.; Denu, J.M.; Garcia, B.A. A novel quantitative mass spectrometry platform for determining protein O-GlcNAcylation Dynamics. Mol. Cell. Proteom. 2016, 15, 2462–2475. [Google Scholar] [CrossRef] [PubMed]
- Haltiwanger, R.S.; Blomberg, M.A.; Hart, G.W. Glycosylation of nuclear and cytoplasmic proteins. Purification and characterization of a uridine diphospho-N-acetylglucosamine: Polypeptide beta-N-acetylglucosaminyltransferase. J. Biol. Chem. 1992, 267, 9005–9013. [Google Scholar] [PubMed]
- Gao, Y.; Wells, L.; Comer, F.I.; Parker, G.J.; Hart, G.W. Dynamic O-Glycosylation of nuclear and cytosolic proteins: Cloning and characterization of a neutral, cytosolic β-N-acetylglucosaminidase from human brain. J. Biol. Chem. 2001, 276, 9838–9845. [Google Scholar] [CrossRef] [PubMed]
- Bond, M.R.; Hanover, J.A. A little sugar goes a long way: The cell biology of O-GlcNAc. J. Cell Biol. 2015, 208, 869–880. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Qian, K. Protein O-GlcNAcylation: Emerging mechanisms and functions. Nat. Rev. Mol. Cell Biol. 2017, 18, 452–465. [Google Scholar] [CrossRef] [PubMed]
- Hardivillé, S.; Hart, G.W. Nutrient regulation of signaling, transcription, and cell physiology by O-GlcNAcylation. Cell Metab. 2014, 20, 208–213. [Google Scholar] [CrossRef] [PubMed]
- Ma, Z.; Vosseller, K. Cancer metabolism and elevated O-GlcNAc in oncogenic signaling. J. Biol. Chem. 2014, 289, 34457–34465. [Google Scholar] [CrossRef] [PubMed]
- Gong, C.-X.; Liu, F.; Iqbal, K. O-GlcNAcylation: A regulator of tau pathology and neurodegeneration. Alzheimer’s Dement. 2016, 12, 1078–1089. [Google Scholar] [CrossRef] [PubMed]
- Vocadlo, D.J.; Hang, H.C.; Kim, E.-J.; Hanover, J.A.; Bertozzi, C.R. A chemical approach for identifying O-GlcNAc-modified proteins in cells. Proc. Natl. Acad. Sci. USA 2003, 100, 9116–9121. [Google Scholar] [CrossRef] [PubMed]
- Zaro, B.W.; Yang, Y.-Y.; Hang, H.C.; Pratt, M.R. Chemical reporters for fluorescent detection and identification of O-GlcNAc-modified proteins reveal glycosylation of the ubiquitin ligase NEDD4-1. Proc. Natl. Acad. Sci. USA 2011, 108, 8146–8151. [Google Scholar] [CrossRef] [PubMed]
- Späte, A.-K.; Schart, V.F.; Häfner, J.; Niederwieser, A.; Mayer, T.U.; Wittmann, V. Expanding the scope of cyclopropene reporters for the detection of metabolically engineered glycoproteins by Diels–Alder reactions. Beilstein J. Org. Chem. 2014, 10, 2235–2242. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yu, S.-H.; Boyce, M.; Wands, A.M.; Bond, M.R.; Bertozzi, C.R.; Kohler, J.J. Metabolic labeling enables selective photocrosslinking of O-GlcNAc-modified proteins to their binding partners. Proc. Natl. Acad. Sci. USA 2012, 109, 4834–4839. [Google Scholar] [CrossRef] [PubMed]
- Boyce, M.; Carrico, I.S.; Ganguli, A.S.; Yu, S.-H.; Hangauer, M.J.; Hubbard, S.C.; Kohler, J.J.; Bertozzi, C.R. Metabolic cross-talk allows labeling of O-linked β-N-acetylglucosamine-modified proteins via the N-acetylgalactosamine salvage pathway. Proc. Natl. Acad. Sci. USA 2011, 108, 3141–3146. [Google Scholar] [CrossRef] [PubMed]
- Chuh, K.N.; Zaro, B.W.; Piller, F.; Piller, V.; Pratt, M.R. Changes in metabolic chemical reporter structure yield a selective probe of O-GlcNAc modification. J. Am. Chem. Soc. 2014, 136, 12283–12295. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Wang, J.; Wen, L.; Zhu, H.; Li, S.; Huang, K.; Jiang, K.; Li, X.; Ma, C.; Qu, J.; et al. An OGA-resistant probe allows specific visualization and accurate identification of O-GlcNAc-modified proteins in cells. ACS Chem. Biol. 2016, 11, 3002–3006. [Google Scholar] [CrossRef] [PubMed]
- Nandi, A.; Sprung, R.; Barma, D.K.; Zhao, Y.; Kim, S.C.; Falck, J.R.; Zhao, Y. Global identification of O-GlcNAc-modified proteins. Anal. Chem. 2006, 78, 452–458. [Google Scholar] [CrossRef] [PubMed]
- Hahne, H.; Sobotzki, N.; Nyberg, T.; Helm, D.; Borodkin, V.S.; van Aalten, D.M.F.; Agnew, B.; Kuster, B. Proteome wide purification and identification of O-GlcNAc-modified proteins using click chemistry and mass spectrometry. J. Proteome Res. 2013, 12, 927–936. [Google Scholar] [CrossRef] [PubMed]
- Hang, H.C.; Yu, C.; Kato, D.L.; Bertozzi, C.R. A metabolic labeling approach toward proteomic analysis of mucin-type O-linked glycosylation. Proc. Natl. Acad. Sci. USA 2003, 100, 14846–14851. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.; Liu, T.-W.; Cecioni, S.; Eskandari, R.; Zandberg, W.F.; Vocadlo, D.J. O-GlcNAc occurs cotranslationally to stabilize nascent polypeptide chains. Nat. Chem. Biol. 2015, 11, 319–325. [Google Scholar] [CrossRef] [PubMed]
- Gurel, Z.; Zaro, B.W.; Pratt, M.R.; Sheibani, N. Identification of O-GlcNAc modification targets in mouse retinal pericytes: Implication of p53 in pathogenesis of diabetic retinopathy. PLoS ONE 2014, 9, e95561. [Google Scholar] [CrossRef] [PubMed]
- Gurel, Z.; Sieg, K.M.; Shallow, K.D.; Sorenson, C.M.; Sheibani, N. Retinal O-linked N-acetylglucosamine protein modifications: Implications for postnatal retinal vascularization and the pathogenesis of diabetic retinopathy. Mol. Vis. 2013, 19, 1047–1059. [Google Scholar] [PubMed]
- Pouilly, S.; Bourgeaux, V.; Piller, F.; Piller, V. Evaluation of analogues of GalNAc as substrates for enzymes of the mammalian galnac salvage pathway. ACS Chem. Biol. 2012, 7, 753–760. [Google Scholar] [CrossRef] [PubMed]
- Kim, E.J. The utilities of chemical reactions and molecular tools for O-GlcNAc proteomic studies. ChemBioChem 2015, 16, 1397–1409. [Google Scholar] [CrossRef] [PubMed]
- Saxon, E.; Bertozzi, C.R. Cell surface engineering by a modified staudinger reaction. Science 2000, 287, 2007–2010. [Google Scholar] [CrossRef] [PubMed]
- Hangauer, M.J.; Bertozzi, C.R. A fret-based fluorogenic phosphine for live-cell imaging with the staudinger ligation. Angew. Chem. Int. Ed. 2008, 47, 2394–2397. [Google Scholar] [CrossRef] [PubMed]
- Prescher, J.A.; Dube, D.H.; Bertozzi, C.R. Chemical remodelling of cell surfaces in living animals. Nature 2004, 430, 873–877. [Google Scholar] [CrossRef] [PubMed]
- Lin, F.L.; Hoyt, H.M.; Van Halbeek, H.; Bergman, R.G.; Bertozzi, C.R. Mechanistic investigation of the staudinger ligation. J. Am. Chem. Soc. 2005, 127, 2686–2695. [Google Scholar] [CrossRef] [PubMed]
- Chang, P.V.; Prescher, J.A.; Hangauer, M.J.; Bertozzi, C.R. Imaging cell surface glycans with bioorthogonal chemical reporters. J. Am. Chem. Soc. 2007, 129, 8400–8401. [Google Scholar] [CrossRef] [PubMed]
- Rostovtsev, V.V.; Green, L.G.; Fokin, V.V.; Sharpless, K.B. A stepwise huisgen cycloaddition process: Copper(i)-catalyzed regioselective “Ligation” of azides and terminal alkynes. Angew. Chem. Int. Ed. 2002, 41, 2596–2599. [Google Scholar] [CrossRef]
- Rodionov, V.O.; Fokin, V.V.; Finn, M.G. Mechanism of the ligand-free CuI-catalyzed Azide-Alkyne cycloaddition reaction. Angew. Chem. Int. Ed. 2005, 44, 2210–2215. [Google Scholar] [CrossRef] [PubMed]
- Presolski, S.I.; Hong, V.; Cho, S.-H.; Finn, M.G. Tailored ligand acceleration of the Cu-catalyzed Azide−Alkyne cycloaddition reaction: Practical and mechanistic implications. J. Am. Chem. Soc. 2010, 132, 14570–14576. [Google Scholar] [CrossRef] [PubMed]
- Agard, N.J.; Baskin, J.M.; Prescher, J.A.; Lo, A.; Bertozzi, C.R. A comparative study of bioorthogonal reactions with azides. ACS Chem. Biol. 2006, 1, 644–648. [Google Scholar] [CrossRef] [PubMed]
- Kennedy, D.C.; McKay, C.S.; Legault, M.C.B.; Danielson, D.C.; Blake, J.A.; Pegoraro, A.F.; Stolow, A.; Mester, Z.; Pezacki, J.P. Cellular consequences of copper complexes used to catalyze bioorthogonal click reactions. J. Am. Chem. Soc. 2011, 133, 17993–18001. [Google Scholar] [CrossRef] [PubMed]
- Agard, N.J.; Prescher, J.A.; Bertozzi, C.R. A strain-promoted [3+2] Azide−Alkyne cycloaddition for covalent modification of biomolecules in living systems. J. Am. Chem. Soc. 2004, 126, 15046–15047. [Google Scholar] [CrossRef] [PubMed]
- Codelli, J.A.; Baskin, J.M.; Agard, N.J.; Bertozzi, C.R. Second-generation difluorinated cyclooctynes for copper-free click chemistry. J. Am. Chem. Soc. 2008, 130, 11486–11493. [Google Scholar] [CrossRef] [PubMed]
- Ning, X.; Guo, J.; Wolfert, M.A.; Boons, G.-J. Visualizing metabolically labeled glycoconjugates of living cells by copper-free and fast huisgen cycloadditions. Angew. Chem. Int. Ed. 2008, 47, 2253–2255. [Google Scholar] [CrossRef] [PubMed]
- Debets, M.F.; van Berkel, S.S.; Schoffelen, S.; Rutjes, F.P.J.T.; van Hest, J.C.M.; van Delft, F.L. Aza-dibenzocyclooctynes for fast and efficient enzyme PEGylation via copper-free (3+2) cycloaddition. Chem. Commun. 2010, 46, 97–99. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jewett, J.C.; Sletten, E.M.; Bertozzi, C.R. Rapid Cu-free click chemistry with readily synthesized biarylazacyclooctynones. J. Am. Chem. Soc. 2010, 132, 3688–3690. [Google Scholar] [CrossRef] [PubMed]
- Sletten, E.M.; Bertozzi, C.R. A hydrophilic azacyclooctyne for Cu-free click chemistry. Org. Lett. 2008, 10, 3097–3099. [Google Scholar] [CrossRef] [PubMed]
- Abdella, P.M.; Smith, P.K.; Royer, G.P. A new cleavable reagent for cross-linking and reversible immobilization of proteins. Biochem. Biophys. Res. Commun. 1979, 87, 734–742. [Google Scholar] [CrossRef]
- Knall, A.-C.; Slugovc, C. Inverse electron demand Diels-Alder (iEDDA)-initiated conjugation: A (high) potential click chemistry scheme. Chem. Soc. Rev. 2013, 42, 5131–5142. [Google Scholar] [CrossRef] [PubMed]
- Niederwieser, A.; Späte, A.-K.; Nguyen, L.D.; Jüngst, C.; Reutter, W.; Wittmann, V. Two-color Glycan labeling of live cells by a combination of diels–alder and click chemistry. Angew. Chem. Int. Ed. 2013, 52, 4265–4268. [Google Scholar] [CrossRef] [PubMed]
- Stairs, S.; Neves, A.A.; Stöckmann, H.; Wainman, Y.A.; Ireland-Zecchini, H.; Brindle, K.M.; Leeper, F.J. Metabolic glycan imaging by isonitrile–tetrazine click chemistry. ChemBioChem 2013, 14, 1063–1067. [Google Scholar] [CrossRef] [PubMed]
- Späte, A.-K.; Bußkamp, H.; Niederwieser, A.; Schart, V.F.; Marx, A.; Wittmann, V. Rapid labeling of metabolically engineered cell-surface glycoconjugates with a carbamate-linked cyclopropene reporter. Bioconjug. Chem. 2014, 25, 147–154. [Google Scholar] [CrossRef] [PubMed]
- Patterson, D.M.; Nazarova, L.A.; Xie, B.; Kamber, D.N.; Prescher, J.A. Functionalized cyclopropenes as bioorthogonal chemical reporters. J. Am. Chem. Soc. 2012, 134, 18638–18643. [Google Scholar] [CrossRef] [PubMed]
- Cole, C.M.; Yang, J.; Šečkutė, J.; Devaraj, N.K. Fluorescent live-cell imaging of metabolically incorporated unnatural cyclopropene-mannosamine derivatives. ChemBioChem 2013, 14, 205–208. [Google Scholar] [CrossRef] [PubMed]
- Blackman, M.L.; Royzen, M.; Fox, J.M. Tetrazine ligation: Fast bioconjugation based on inverse-electron-demand diels−alder reactivity. J. Am. Chem. Soc. 2008, 130, 13518–13519. [Google Scholar] [CrossRef] [PubMed]
- Devaraj, N.K.; Weissleder, R.; Hilderbrand, S.A. Tetrazine-based cycloadditions: Application to pretargeted live cell imaging. Bioconjug. Chem. 2008, 19, 2297–2299. [Google Scholar] [CrossRef] [PubMed]
- Taylor, M.T.; Blackman, M.L.; Dmitrenko, O.; Fox, J.M. Design and synthesis of highly reactive dienophiles for the tetrazine–trans-cyclooctene ligation. J. Am. Chem. Soc. 2011, 133, 9646–9649. [Google Scholar] [CrossRef] [PubMed]
- Devaraj, N.K.; Hilderbrand, S.; Upadhyay, R.; Mazitschek, R.; Weissleder, R. Bioorthogonal turn-on probes for imaging small molecules inside living cells. Angew. Chem. Int. Ed. 2010, 49, 2869–2872. [Google Scholar] [CrossRef] [PubMed]
- Lang, K.; Davis, L.; Wallace, S.; Mahesh, M.; Cox, D.J.; Blackman, M.L.; Fox, J.M.; Chin, J.W. Genetic encoding of bicyclononynes and trans-cyclooctenes for site-specific protein labeling in vitro and in live mammalian cells via rapid fluorogenic diels–alder reactions. J. Am. Chem. Soc. 2012, 134, 10317–10320. [Google Scholar] [CrossRef] [PubMed]
- Collins, B.E.; Paulson, J.C. Cell surface biology mediated by low affinity multivalent protein–glycan interactions. Curr. Opin. Chem. Biol. 2004, 8, 617–625. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez, A.C.; Yu, S.-H.; Li, B.; Zegzouti, H.; Kohler, J.J. Enhanced transfer of a photocross-linking N-acetylglucosamine (GlcNAc) analog by an O-GlcNAc transferase mutant with converted substrate specificity. J. Biol. Chem. 2015, 290, 22638–22648. [Google Scholar] [CrossRef] [PubMed]
- Mahal, L.K.; Yarema, K.J.; Bertozzi, C.R. Engineering chemical reactivity on cell surfaces through oligosaccharide biosynthesis. Science 1997, 276, 1125–1128. [Google Scholar] [CrossRef] [PubMed]
- Nauman, D.A.; Bertozzi, C.R. Kinetic parameters for small-molecule drug delivery by covalent cell surface targeting. Biochim. Biophys. Acta 2001, 1568, 147–154. [Google Scholar] [CrossRef]
- Klement, E.; Lipinszki, Z.; Kupihár, Z.; Udvardy, A.; Medzihradszky, K.F. Enrichment of O-GlcNAc modified proteins by the periodate oxidation−hydrazide resin capture approach. J. Proteome Res. 2010, 9, 2200–2206. [Google Scholar] [CrossRef] [PubMed]
- Khidekel, N.; Ficarro, S.B.; Peters, E.C.; Hsieh-Wilson, L.C. Exploring the O-GlcNAc proteome: Direct identification of O-GlcNAc-modified proteins from the brain. Proc. Natl. Acad. Sci. USA 2004, 101, 13132–13137. [Google Scholar] [CrossRef] [PubMed]
- Khidekel, N.; Ficarro, S.B.; Clark, P.M.; Bryan, M.C.; Swaney, D.L.; Rexach, J.E.; Sun, Y.E.; Coon, J.J.; Peters, E.C.; Hsieh-Wilson, L.C. Probing the dynamics of O-GlcNAc glycosylation in the brain using quantitative proteomics. Nat. Chem. Biol. 2007, 3, 339–348. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Park, K.; Comer, F.; Hsieh-Wilson, L.C.; Saudek, C.D.; Hart, G.W. Site-specific GlcNAcylation of human erythrocyte proteins: Potential biomark(s) for diabetes. Diabetes 2009, 58, 309–317. [Google Scholar] [CrossRef] [PubMed]
- Jencks, W.P. Studies on the mechanism of oxime and semicarbazone formation1. J. Am. Chem. Soc. 1959, 81, 475–481. [Google Scholar] [CrossRef]
- Sander, E.G.; Jencks, W.P. Equilibria for additions to the carbonyl group. J. Am. Chem. Soc. 1968, 90, 6154–6162. [Google Scholar] [CrossRef]
- Clark, P.M.; Dweck, J.F.; Mason, D.E.; Hart, C.R.; Buck, S.B.; Peters, E.C.; Agnew, B.J.; Hsieh-Wilson, L.C. Direct In-Gel fluorescence detection and cellular imaging of O-GlcNAc-modified proteins. J. Am. Chem. Soc. 2008, 130, 11576–11577. [Google Scholar] [CrossRef] [PubMed]
- Parker, B.L.; Gupta, P.; Cordwell, S.J.; Larsen, M.R.; Palmisano, G. Purification and identification of O-GlcNAc-modified peptides using phosphate-based alkyne click chemistry in combination with titanium dioxide chromatography and mass spectrometry. J. Proteome Res. 2011, 10, 1449–1458. [Google Scholar] [CrossRef] [PubMed]
- Sakabe, K.; Wang, Z.; Hart, G.W. β-N-Acetylglucosamine (O-GlcNAc) is part of the histone code. Proc. Natl. Acad. Sci. USA 2010, 107, 19915–19920. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Udeshi, N.D.; Slawson, C.; Compton, P.D.; Sakabe, K.; Cheung, W.D.; Shabanowitz, J.; Hunt, D.F.; Hart, G.W. Extensive crosstalk between O-GlcNAcylation and phosphorylation regulates cytokinesis. Sci. Signal. 2010, 3, ra2. [Google Scholar] [CrossRef] [PubMed]
- Alfaro, J.F.; Gong, C.-X.; Monroe, M.E.; Aldrich, J.T.; Clauss, T.R.W.; Purvine, S.O.; Wang, Z.; Camp, D.G.; Shabanowitz, J.; Stanley, P.; et al. Tandem mass spectrometry identifies many mouse brain O-GlcNAcylated proteins including EGF domain-specific O-GlcNAc transferase targets. Proc. Natl. Acad. Sci. USA 2012, 109, 7280–7285. [Google Scholar] [CrossRef] [PubMed]
- Sola-Penna, M.; Da Silva, D.; Coelho, W.S.; MarinhO-Carvalho, M.M.; Zancan, P. Regulation of mammalian muscle type 6-phosphofructo-1-kinase and its implication for the control of the metabolism. IUBMB Life 2010, 62, 791–796. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yi, W.; Clark, P.M.; Mason, D.E.; Keenan, M.C.; Hill, C.; Goddard, W.A.; Peters, E.C.; Driggers, E.M.; Hsieh-Wilson, L.C. Phosphofructokinase 1 glycosylation regulates cell growth and metabolism. Science 2012, 337, 975–980. [Google Scholar] [CrossRef] [PubMed]
- Robinson, P.V.; Tsai, C.-T.; de Groot, A.E.; McKechnie, J.L.; Bertozzi, C.R. Glyco-seek: Ultrasensitive detection of protein-specific glycosylation by proximity ligation polymerase chain reaction. J. Am. Chem. Soc. 2016, 138, 10722–10725. [Google Scholar] [CrossRef] [PubMed]
- Ramakrishnan, P.; Clark, P.M.; Mason, D.E.; Peters, E.C.; Hsieh-Wilson, L.C.; Baltimore, D. Activation of the transcriptional function of the NF-κB protein c-rel by O-GlcNAc glycosylation. Sci. Signal. 2013, 6, ra75. [Google Scholar] [CrossRef] [PubMed]
- Torres, C.R.; Hart, G.W. Topography and polypeptide distribution of terminal N-acetylglucosamine residues on the surfaces of intact lymphocytes evidence for O-linked GlcNAc. J. Biol. Chem. 1984, 259, 3308–3317. [Google Scholar] [PubMed]
- Banda, K.; Gregg, C.J.; Chow, R.; Varki, N.M.; Varki, A. Metabolism of vertebrate amino sugars with n-glycolyl groups: Mechanisms underlying gastrointestinal incorporation of the non-human sialic acid xeno-autoantigen n-glycolylneuraminic acid. J. Biol. Chem. 2012, 287, 28852–28864. [Google Scholar] [CrossRef] [PubMed]






© 2018 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, E.J. Chemical Reporters and Their Bioorthogonal Reactions for Labeling Protein O-GlcNAcylation. Molecules 2018, 23, 2411. https://doi.org/10.3390/molecules23102411
Kim EJ. Chemical Reporters and Their Bioorthogonal Reactions for Labeling Protein O-GlcNAcylation. Molecules. 2018; 23(10):2411. https://doi.org/10.3390/molecules23102411
Chicago/Turabian StyleKim, Eun Ju. 2018. "Chemical Reporters and Their Bioorthogonal Reactions for Labeling Protein O-GlcNAcylation" Molecules 23, no. 10: 2411. https://doi.org/10.3390/molecules23102411
APA StyleKim, E. J. (2018). Chemical Reporters and Their Bioorthogonal Reactions for Labeling Protein O-GlcNAcylation. Molecules, 23(10), 2411. https://doi.org/10.3390/molecules23102411