Identification and Pharmacokinetic Studies on Complanatuside and Its Major Metabolites in Rats by UHPLC-Q-TOF-MS/MS and LC-MS/MS
Abstract
:1. Introduction
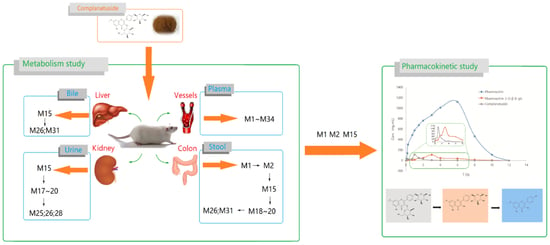
2. Results
2.1. Metabolites Study
2.1.1. Fragmentation Studies of Complanatuside Standard
2.1.2. UHPLC–Q-TOF-MS/MS Analysis
2.1.3. Parent Drug and Metabolites of the Parent Drug
Parent compound M1
Metabolites M2 and M15
2.1.4. Metabolites of the Degradation Products
Methylated metabolites (M3; M16)
Demethylated metabolites (M4; M17)
Demethoxy metabolites (M5; M6; M7; M18)
Hydroxylated metabolites (M8; M9; M19; M20; M21; M22)
Sulfonated metabolites (M10; M23; M24; M25; M26; M27; M28; M29; M30)
Glucuronidated metabolites (M11; M12; M13; M14; M31; M32; M33)
Dehydrated metabolites (M34)
2.2. Pharmacokinetic Study
2.2.1. Selection of Internal Standard
2.2.2. Method Validation
Selectivity and Carryover
Linearity and Sensitivity
Recovery and Matrix Effect
Stability
Precision and Accuracy
2.2.3. Pharmacokinetics of Complanatuside and Two Metabolites
3. Discussion
4. Materials and Methods
4.1. Chemicals and Reagents
4.2. Animal Experiments
4.3. Sample Preparation
4.3.1. Metabolism Study
4.3.2. Pharmacokinetic Study
4.4. Instruments and Experimental Conditions
4.4.1. UHPLC–Q-TOF-MS Conditions
4.4.2. LC-MS/MS Conditions
4.5. Method Validation
4.6. Data Analysis of the Pharmacokinetic Study
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of interest
References
- Xu, P.; Zhang, W.B.; Cai, X.H.; Qiu, P.Y.; Hao, M.H.; Lu, D.D. Activating AKT to inhibit JNK by troxerutin antagonizes radiation-induced PTEN activation. Eur. J. Pharmacol. 2017, 795, 66–74. [Google Scholar] [CrossRef] [PubMed]
- Kalita, B.; Ranjan, R.; Singh, A.; Yashavarddhan, M.H.; Bajaj, S.; Gupta, M.L. A Combination of Podophyllotoxin and Rutin Attenuates Radiation Induced Gastrointestinal Injury by Negatively Regulating NF-κB/p53 Signaling in Lethally Irradiated Mice. PLoS ONE 2016, 11, e0168525. [Google Scholar] [CrossRef] [PubMed]
- Hafez, M.M.; Al-Harbi, N.O.; Al-Hoshani, A.R.; Al-Hosaini, K.A.; Al Shrari, S.D.; Al Rejaie, S.S.; Sayed-Ahmed, M.M.; Al-Shabanah, O.A. Hepato-protective effect of rutin via IL-6/STAT3 pathway in CCl4-induced hepatotoxicity in rats. Biol. Res. 2015, 48, 30. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.C.; Byun, S.H.; Yang, C.H.; Kim, C.Y.; Kim, J.W.; Kim, S.G. Cytoprotective effects of Glycyrrhizae radix extract and its active component liquiritigenin against cadmium-induced toxicity (effects on bad translocation and cytochrome c-mediated PARP cleavage). Toxicology 2004, 197, 239–251. [Google Scholar] [CrossRef] [PubMed]
- Hou, Z.; Hu, Y.; Yang, X.; Chen, W. Antihypertensive effects of Tartary buckwheat flavonoids by improvement of vascular insulin sensitivity in spontaneously hypertensive rats. Food Funct. 2017, 8, 4217–4228. [Google Scholar] [CrossRef] [PubMed]
- Dower, J.I.; Geleijnse, J.M.; Gijsbers, L.; Schalkwijk, C.; Kromhout, D.; Hollman, P.C. Supplementation of the Pure Flavonoids Epicatechin and Quercetin Affects Some Biomarkers of Endothelial Dysfunction and Inflammation in (Pre)Hypertensive Adults: A Randomized Double-Blind, Placebo-Controlled, Crossover Trial. J. Nutr. 2015, 145, 1459–1463. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jadoon, S.; Karim, S.; Bin Asad, M.H.H.; Akram, M.R.; Khan, A.K.; Malik, A.; Chen, C.Y.; Murtaza, G. Anti-Aging Potential of Phytoextract Loaded-Pharmaceutical Creams for Human Skin Cell Longetivity. Oxid. Med. Cell Longev. 2015, 709628. [Google Scholar] [CrossRef]
- Lephart, E.D. Skin aging and oxidative stress: Equol’s anti-aging effects via biochemical and molecular mechanisms. Ageing Res. Rev. 2016, 31, 36–54. [Google Scholar] [CrossRef]
- Irakli, M.N.; Samanidou, V.F.; Biliaderis, C.G.; Papadoyannis, I.N. Simultaneous determination of phenolic acids and flavonoids in rice using solid-phase extraction and RP-HPLC with photodiode array detection. J. Sep. Sci. 2012, 35, 1603–1611. [Google Scholar] [CrossRef]
- Qiu, L.; Jiao, Y.; Xie, J.Z.; Huang, G.K.; Qiu, S.L.; Miao, J.H.; Yao, X.S. Five new flavonoid glycosides from Nerviliafordii. J. Asian Nat. Prod. Res. 2013, 15, 589–599. [Google Scholar] [CrossRef]
- Wiczkowski, W.; Romaszko, J.; Bucinski, A.; Szawara-Nowak, D.; Honke, J.; Zielinski, H.; Piskula, M.K. Quercetin from shallots (Allium cepa L. var. aggregatum) is more bioavailable than its glucosides. J. Nutr. 2008, 138, 885–888. [Google Scholar] [PubMed]
- Terao, J. Factors modulating bioavailability of quercetin-related flavonoids and the consequences of their vascular function. Biochem. Pharmacol. 2017, 139, 15–23. [Google Scholar] [CrossRef] [PubMed]
- Hu, S.C.; Hong, K.; Song, Y.C.; Liu, J.Y.; Tan, R.X. Biotransformation of soybean isoflavones by a marine Streptomyces sp 060524 and cytotoxicity of the products. World J. Microb. Biot. 2009, 25, 115–121. [Google Scholar] [CrossRef]
- Teng, J.; Li, Y.L.; Yu, W.Z.; Zhao, Y.L.; Hu, X.Q.; Tao, N.P.; Wang, M.F. Naringenin, a common flavanone, inhibits the formation of AGEs in bread and attenuates AGEs-induced oxidative stress and inflammation in RAW264.7 cells. Food Chem. 2018, 269, 35–42. [Google Scholar] [CrossRef] [PubMed]
- Zhang, G.; Chen, S.S.; Zhou, W.; Meng, J.; Deng, K.; Zhou, H.N.; Hu, N.; Suo, Y.R. Rapid qualitative and quantitative analyses of eighteen phenolic compounds from Lyciumruthenicum Murray by UPLC-Q-Orbitrap MS and their antioxidant activity. Food Chem. 2018, 269, 150–156. [Google Scholar] [CrossRef] [PubMed]
- Fernandes, I.; Perez-Gregorio, R.; Soares, S.; Mateus, N.; Freitas, V. Wine Flavonoids in Health and Disease Prevention. Molecules 2017, 22, 292. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Wang, A.Q.; Li, X.J.; Fan, X.; Yin, S.S.; Lan, K. A chemical profiling strategy for semi-quantitative analysis of flavonoids in Ginkgo extracts. J. Pharm. Biomed. Anal. 2016, 123, 147–154. [Google Scholar] [CrossRef]
- Zhang, J.J.; Yan, X.L.; Zhang, Y.J.; Wang, Y.P.; Zhang, Q.J.; Zeng, F.Y.; Li, W.; Ou, L.N. Determination of complanatoside A in semen Astragali complanati by HPLC. China J. Chin. Mater. Med. 2005, 30, 600–602. [Google Scholar]
- National Pharmacopoeia Committee. Pharmacopoeia of People’s Republic of China; China Medical Science and Technology Press: Beijing, China, 2010; pp. 181–182. [Google Scholar]
- Li, N.; Liu, Y.; Cao, Y.C.; Wei, Z.X.; Pang, L.; Wang, J.M. Quantification of complanatoside A in rat plasma using LC-MS/MS and its application to a pharmacokinetic study. Biomed. Chromatogr. 2016, 30, 888–893. [Google Scholar] [CrossRef]
- Fang, S.H.; Rao, Y.K.; Tzeng, Y.M. Anti-oxidant and inflammatory mediator’s growth inhibitory effects of compounds isolated from Phyllanthusurinaria. J. Ethnopharmacol. 2008, 116, 333–340. [Google Scholar] [CrossRef]
- Zhoua, X.F.; Tong, G.T.; Wang, X.W.; He, Y. Anti-proliferative constituents from Selaginellamoellendorffii. Nat. Prod. Commun. 2016, 11, 623–626. [Google Scholar] [PubMed]
- Miao, X.; Wang, J.; Chen, L.; Peng, Z.; Chen, Y. Identification of in vivo and in vitro metabolites of 4,5-dimethoxycanthin-6-one by HPLC-Q-TOF-MS/MS. J. Chromatogr. B Analyt. Technol. Biomed. Life. Sci. 2016, 1020, 78–84. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Wang, C.; Li, H.; Yu, T.; Tan, L. Simultaneous Determination of Formononetin, Calycosin and Rhamnocitrin from Astragalus Complanatus by UHPLC-MS-MS in Rat Plasma: Application to a Pharmacokinetic Study. J. Chromatogr. Sci. 2016, 54, 1605–1612. [Google Scholar] [CrossRef] [PubMed]
- Lin, P.; Qin, Z.; Yao, Z.; Wang, L.; Zhang, W.; Yu, Y.; Dai, Y.; Zhou, H.; Yao, X. Metabolites profile of GualouXiebaiBaijiu decoction (a classical traditional Chinese medicine prescription) in rats by ultra-performance liquid chromatography coupled with quadrupole time-of-flight tandem mass spectrometry. J. Chromatogr. B Analyt. Technol. Biomed. Life. Sci. 2018, 1085, 72–88. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Zheng, S.; Li, L.; Jiang, H. Metabolism of flavonoids in human: A comprehensive review. Curr. Drug. Metab. 2014, 15, 48–61. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Zhu, C.C.; Zhao, Z.X.; Lin, C.Z. Simultaneous determination of seven flavonoids in Nerviliafordii with HPLC. Acta. Pharm. Sin. 2011, 46, 1237–1240. [Google Scholar]
Sample Availability: Samples of the compounds are not available from the authors. |







| No. | Time (min) | Formula (M − H/+HCOO) | Experimental (Da) | Fragment Ion (Da) | Metabolite Description | Error (ppm) | Samples | |
|---|---|---|---|---|---|---|---|---|
| M1 * | 0.79 | C28H32O16 | 669.1663 | 461.1089 | 299.0553 | Parent | 0.62 | P, S * |
| M2 | 0.82 | C22H22O11 | 461.1089 | 299.0558 | 283.0236 | Deglycosylation | 1.20 | P, S |
| M3 | 3.20 | C23H24O11 | 475.1246 | 429.2995 | M2 methylation | 1.18 | P | |
| M4 | 2.20 | C21H20O11 | 447.0929 | 429.2978 | 149.0019 | M2 demethylation | 0.36 | P |
| M5 | 1.04 | C21H20O10 | 431.0983 | 299.1995 | 148.9996 | M2 demethylation | 1.11 | P |
| M6 | 2.19 | C22H22O10 | 445.1140 | 296.9965 | M2 demethylation | 1.18 | P | |
| M7 | 2.46 | C20H18O10 | 463.0871 | 391.0617 | 152.9916 | M2 demethylation | 1.32 | P |
| M8 | 3.99 | C21H20O12 | 463.0882 | 431.1391 | 355.1693 | M2 hydroxylation | 1.18 | P |
| M9 | 1.23 | C20H18O11 | 479.0825 | 391.2848 | 227.2567 | M2 hydroxylation | 0.15 | P |
| M10 | 1.58 | C22H22O15S | 557.0607 | 255.2311 | 181.9966 | M2 sulfonation | 1.05 | P |
| M11 | 9.20 | C27H28O16 | 653.1348 | 421.3522 | 175.0235 | M2 glucuronidation | 0.97 | P |
| M12 | 1.14 | C28H30O18 | 653.1359 | 447.1223 | 335.2226 | M2 glucuronidation | 0.78 | P |
| M13 | 9.23 | C29H32O18 | 713.1560 | 653.3056 | 447.1289 | M2 glucuronidation | 0.78 | P |
| M14 | 4.12 | C29H32O20S | 731.1135 | 317.2127 | 299.1980 | M2 glucuronidation | 0.77 | P |
| M15 | 1.08 | C16H12O6 | 299.0559 | 285.0312 | 271.0606 | Deglycosylation of M2 | 1.12 | P, B *,S, U * |
| M16 | 0.92 | C17H14O6 | 313.0718 | 285.1236 | 269.1281 | M15 methylation | 1.87 | P |
| M17 | 1.09 | C15H10O6 | 331.0460 | 269.1902 | 149.0965 | M15 demethylation | 2.12 | P, U |
| M18 | 1.35 | C15H10O5 | 269.0449 | 241.0491 | 225.0540 | M15 demethoxylation | 0.36 | P, S, U |
| M19 | 1.28 | C16H12O7 | 315.0510 | 300.0264 | 165.0183 | M15 hydroxylation | 1.65 | P, S, U |
| M20 | 0.82 | C17H14O7 | 375.0711 | 316.1675 | 285.1854 | M15 hydroxylation | 1.66 | P, S, U |
| M21 | 0.91 | C15H10O7 | 347.0398 | 285.2577 | 135.1503 | M15 hydroxylation | 1.68 | P |
| M22 | 0.78 | C14H8O6 | 271.0248 | 255.0763 | M15 hydroxylation | 2.01 | P | |
| M23 | 2.86 | C16H12O9S | 379.0129 | 299.0564 | 271.0616 | M15 sulfonation | 1.37 | P |
| M24 | 0.76 | C17H14O9S | 393.0315 | 313.1804 | 299.1640 | M15 sulfonation | 1.39 | P |
| M25 | 0.77 | C15H10O9S | 411.0017 | 287.1634 | M15 sulfonation | 1.49 | P, U | |
| M26 | 0.63 | C15H10O8S | 349.0024 | 331.2625 | M15 sulfonation | 1.56 | P, B, S, U | |
| M27 | 0.90 | C16H12O8S | 363.0180 | 285.0581 | 257.1902 | M15 sulfonation | 1.47 | P |
| M28 | 1.17 | C14H8O8S | 334.9845 | 255.1022 | 240.0785 | M15 sulfonation | 4.95 | P, S, U |
| M29 | 0.76 | C16H12O10S | 395.0078 | 347.2233 | 331.2281 | M15 sulfonation | 1.38 | P |
| M30 | 1.17 | C15H10O10S | 380.9922 | 195.1384 | 181.1232 | M15 sulfonation | 1.43 | P |
| M31 | 0.81 | C22H20O12 | 475.0877 | 299.0553 | 284.0332 | M15 glucuronidation | 0.10 | P, B |
| M32 | 3.36 | C23H22O13 | 505.0988 | 447.1352 | M15 glucuronidation | 1.15 | P | |
| M33 | 4.07 | C20H16O11 | 431.0620 | 355.1686 | M15 glucuronidation | 1.30 | P | |
| M34 | 1.21 | C28H32O15 | 653.1761 | 447.1342 | 285.1258 | M1 dehydration | 7.11 | P |
| Analytes | Linear Equation | R2 | Linear Range (ng/mL) | LLOQ (ng/mL) |
|---|---|---|---|---|
| CPS | Y = 0.0137X + 0.2118 | 0.9975 | 5.20–520.00 | 5.20 |
| RNG | Y = 0.2714X − 0.4355 | 0.9951 | 2.04–510.00 | 2.04 |
| RNC | Y = 0.2656X + 1.1438 | 0.9948 | 1.02–510.00 | 1.02 |
| Analysts | QC Concentration (ng/mL) | Extraction Recovery | Matrix Effect | ||
|---|---|---|---|---|---|
| Accuracy (%) | RSD (%) | Accuracy (%) | RSD (%) | ||
| CPS | 20 | 73.5 ± 8.7 | 5.5 | 102.1 ± 4.4 | 11.3 |
| 125 | 81.0 ± 5.1 | 6.0 | 92.7 ± 14.6 | 8.3 | |
| 500 | 76.7 ± 4.3 | 6.4 | 98.7 ± 2.9 | 7.4 | |
| RNG | 20 | 83.7 ± 2.8 | 12.6 | 86.0 ± 7.5 | 5.4 |
| 125 | 83.3 ± 1.9 | 6.7 | 97.7 ± 2.1 | 5.9 | |
| 500 | 83.2 ± 5.1 | 6.1 | 98.2 ± 2.9 | 3.8 | |
| RNC | 20 | 90.3 ± 3.7 | 7.5 | 101.1 ± 4.3 | 4.8 |
| 125 | 89.0 ± 5.5 | 6.4 | 93.2 ± 2.3 | 3.3 | |
| 500 | 88.7 ± 3.7 | 9.8 | 95.6 ± 4.8 | 2.4 | |
| Analytes | QC Concentration (ng/mL) | Post Preparation Stability | Short-Term Stability | Long-Term Stability | Freeze-Thaw Stability | ||||
|---|---|---|---|---|---|---|---|---|---|
| Mean (ng/mL) | RSD (%) | Mean (ng/mL) | RSD (%) | Mean (ng/mL) | RSD (%) | Mean (ng/mL) | RSD (%) | ||
| CPS | 20 | 18.6 ± 1.3 | 7.0 | 18.5 ± 1.4 | 7.6 | 17.4 ± 1.3 | 7.5 | 17.4 ± 1.2 | 6.9 |
| 125 | 124.4 ± 11.2 | 9.0 | 124.1 ± 6.2 | 5.0 | 124.2 ± 9.1 | 7.3 | 122.1 ± 5.5 | 4.5 | |
| 500 | 489.3 ± 22.6 | 4.6 | 469.2 ± 12.5 | 2.7 | 486.5 ± 26.2 | 5.4 | 487.3 ± 23.7 | 4.9 | |
| RNG | 20 | 19.3 ± 1.4 | 7.3 | 19.2 ± 1.3 | 6.8 | 18.7 ± 1.3 | 7.0 | 18.8 ± 0.9 | 4.8 |
| 125 | 124.7 ± 10.9 | 8.7 | 123.6 ± 6.7 | 5.4 | 123.4 ± 10.6 | 8.6 | 123.5 ± 7.6 | 6.2 | |
| 500 | 491.5 ± 23.4 | 4.8 | 486.9 ± 23.3 | 4.8 | 479.4 ± 24.8 | 5.2 | 479.7 ± 12.8 | 2.7 | |
| RNC | 20 | 19.2 ± 1.2 | 6.3 | 19.0 ± 1.1 | 5.8 | 19.7 ± 1.1 | 5.6 | 19.1 ± 0.8 | 4.1 |
| 125 | 124.1 ± 10.5 | 8.5 | 124.0 ± 5.9 | 4.6 | 125.5 ± 6.9 | 5.5 | 124.7 ± 9.7 | 7.8 | |
| 500 | 476.3 ± 33.6 | 7.1 | 473.1 ± 25.1 | 5.3 | 495.3 ± 20.5 | 4.1 | 482.3 ± 14.3 | 3.1 | |
| Analytes | QC Concentration (ng/mL) | Intra-Day | Inter-Day | ||||
|---|---|---|---|---|---|---|---|
| Actual Conc. (ng/mL) | Precision (RSD, %) | Accuracy (RE, %) | Actual Conc. (ng/mL) | Precision (RSD, %) | Accuracy (RE, %) | ||
| CPS | 20 | 19.6 ± 0.6 | 3.1 | 2.0 | 19.8 ± 1.9 | 9.6 | 1.0 |
| 125 | 124.8 ± 7.3 | 5.8 | 0.2 | 124.8 ± 6.2 | 5.0 | 0.2 | |
| 500 | 504.7 ± 24.2 | 4.8 | −0.9 | 494.8 ± 17.9 | 3.6 | 1.0 | |
| RNG | 20 | 19.2 ± 1.1 | 5.7 | 4.0 | 18.8 ± 1.7 | 9.0 | 6.0 |
| 125 | 123.2 ± 5.8 | 4.7 | 1.4 | 125.4 ± 2.19 | 1.7 | −0.3 | |
| 500 | 504.7 ± 21.4 | 4.2 | −0.9 | 498.6 ± 18.2 | 3.7 | 0.3 | |
| RNC | 20 | 19.3 ± 1.0 | 5.2 | 3.5 | 18.7 ± 1.9 | 10.1 | 6.5 |
| 125 | 125.8 ± 4.9 | 3.9 | −0.6 | 122.4 ± 10.5 | 8.6 | 2.1 | |
| 500 | 496.9 ± 13.6 | 2.7 | 0.6 | 505.8 ± 13.5 | 2.7 | −1.2 | |
| Analytes | Cmax (ng/mL) | Tmax (h) | t1/2 (h) | AUC(0-t) (µg/L·h) | AUC(0-∞) (µg/L·h) | MRT(0-t) (h) |
|---|---|---|---|---|---|---|
| CPS | 119.15 ± 11.25 | 1.00 ± 0.36 | 0.51 ± 0.04 | 143.52 ± 15.73 | 143.52 ± 15.73 | 1.72 ± 0.19 |
| RNG | 111.64 ± 14.68 | 3.00 ± 0.25 | 1.33 ± 0.55 | 381.73 ± 24.13 | 387.21 ± 28.06 | 4.28 ± 0.55 |
| RNC | 1122.18 ± 113.32 | 5.33 ± 0.63 | 4.15 ± 0.49 | 6540.14 ± 433.70 | 6627.61 ± 471.83 | 4.99 ± 0.11 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yao, Y.-F.; Lin, C.-Z.; Liu, F.-L.; Zhang, R.-J.; Zhang, Q.-Y.; Huang, T.; Zou, Y.-S.; Wang, M.-Q.; Zhu, C.-C. Identification and Pharmacokinetic Studies on Complanatuside and Its Major Metabolites in Rats by UHPLC-Q-TOF-MS/MS and LC-MS/MS. Molecules 2019, 24, 71. https://doi.org/10.3390/molecules24010071
Yao Y-F, Lin C-Z, Liu F-L, Zhang R-J, Zhang Q-Y, Huang T, Zou Y-S, Wang M-Q, Zhu C-C. Identification and Pharmacokinetic Studies on Complanatuside and Its Major Metabolites in Rats by UHPLC-Q-TOF-MS/MS and LC-MS/MS. Molecules. 2019; 24(1):71. https://doi.org/10.3390/molecules24010071
Chicago/Turabian StyleYao, Yu-Feng, Chao-Zhan Lin, Fang-Le Liu, Run-Jing Zhang, Qiu-Yu Zhang, Tao Huang, Yuan-Sheng Zou, Mei-Qi Wang, and Chen-Chen Zhu. 2019. "Identification and Pharmacokinetic Studies on Complanatuside and Its Major Metabolites in Rats by UHPLC-Q-TOF-MS/MS and LC-MS/MS" Molecules 24, no. 1: 71. https://doi.org/10.3390/molecules24010071
APA StyleYao, Y. -F., Lin, C. -Z., Liu, F. -L., Zhang, R. -J., Zhang, Q. -Y., Huang, T., Zou, Y. -S., Wang, M. -Q., & Zhu, C. -C. (2019). Identification and Pharmacokinetic Studies on Complanatuside and Its Major Metabolites in Rats by UHPLC-Q-TOF-MS/MS and LC-MS/MS. Molecules, 24(1), 71. https://doi.org/10.3390/molecules24010071