High Production of Chitinolytic Activity in Halophilic Conditions by a New Marine Strain of Clonostachys rosea
Abstract
:1. Introduction
2. Results and Discussion
2.1. Plate Screening for Chitinolytic Enzyme Producers
2.2. Secondary Screening for Chitinolytic Enzymes
2.3. Optimal Growth Conditions with Respect to Temperature, pH and Salinity
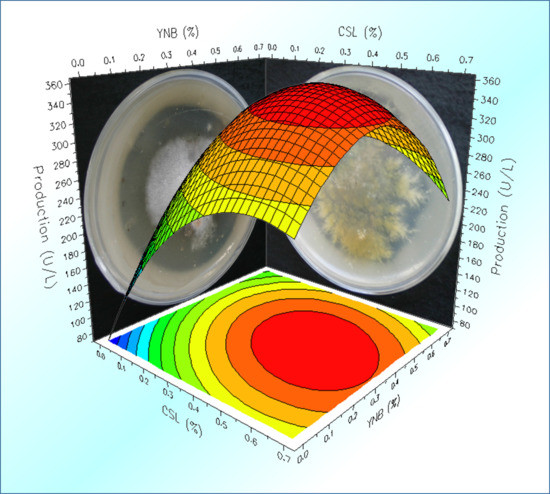
2.4. Optimization of the Cultural Medium by Response Surface Methodology
3. Materials and Methods
3.1. Collection of Samples and Isolation of Pure Cultures of Fungi
3.2. Strain Identification
3.3. Screening Procedure
3.4. Optimization of Growth Conditions of pH, Temperature and Salinity
3.5. Optimization of Culture Medium by RSM Factorial Design
3.6. Analytical Methods
4. Conclusions
Author Contributions
Acknowledgments
Conflicts of Interest
References
- Fenice, M.; Gallo, A.M.; Juarez-Jimenez, B.; Gonzalez-Lopez, J. Screening for extracellular enzyme activities by bacteria isolated from samples collected in the Tyrrhenian Sea. Ann. Microbiol. 2007, 57, 93–99. [Google Scholar] [CrossRef]
- Trincone, A. Marine biocatalysts: enzymatic features and applications. Mar. Drugs 2011, 9, 478–499. [Google Scholar] [CrossRef] [PubMed]
- Bonugli-Santos, R.C.; Dos Santos Vasconcelos, M.R.; Passarini, M.R.; Vieira, G.A.; Lopes, V.C.; Mainardi, P.H.; Feitosa, V.A. Marine-derived fungi: diversity of enzymes and biotechnological applications. Front. Microbiol. 2015, 6, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Srivastava, A.; Srivastava, M. Enzymes Market Type (Protease, Carbohydrase, Lipase, Polymerase and Nuclease, and Other Types), Source (Microorganisms, Plants, and Animals), Reaction Type (Hydrolase, Oxidoreductase, Transferase, Lyase, and Other Reaction Types), and Application (Food and Beverages, Household Care, Bioenergy, Pharmaceutical and Biotechnology, Feed, and Other Applications)—Global Opportunity Analysis and Industry Forecast, 2017–2024. 2018. Available online: https://www.alliedmarketresearch.com/enzymes-market (accessed on May 2018).
- Alma’abadi, A.D.; Gojobori, T.; Mineta, K. Marine metagenome as a resource for novel enzymes. Genom. Proteom. Bioinform. 2015, 13, 290–295. [Google Scholar]
- Debashish, G.; Malay, S.; Barindra, S.; Joydeep, M. Marine Enzymes. In Marine Biotechnology I. Advances in Biochemical Engineering/Biotechnology; Ulber, R., Le Gal, Y., Eds.; Springer: Heidelberg/Berlin, Germany, 2005. [Google Scholar]
- Atalla, M.M.; Zeinab, H.K.; Eman, R.H.; Amani, A.Y.; Abeer, A.A.E.A. Screening of some marine-derived fungal isolates for lignin degrading enzymes (LDEs) production. Agric. Biol. J. N. Am. 2010, 1, 591–599. [Google Scholar]
- Thirunavukkarasu, N.; Jahnes, B.; Broadstock, A.; Rajulu, M.G.; Murali, T.S.; Gopalan, V.; Suryanarayanan, T.S. Screening marine-derived endophytic fungi for xylan-degrading enzymes. Curr. Sci. 2015, 112–120. [Google Scholar]
- Fenice, M. The psychrotolerant Antarctic fungus Lecanicillium muscarium CCFEE 5003: A powerful producer of cold-tolerant chitinolytic enzymes. Molecules 2016, 21, 447. [Google Scholar] [CrossRef]
- Poulicek, M.; Machiroux, R.; Toussaint, C. Chitin diagenesis in deep-water sediments. In Chitin in Nature and Technology; Springer: Boston, MA, USA, 1986; pp. 523–530. [Google Scholar]
- Keyhani, N.O.; Roseman, S. Physiological aspects of chitin catabolism in marine bacteria. Biochim. Biophys. Acta Gen. Subj. 1999, 1473, 108–122. [Google Scholar] [CrossRef]
- Souza, C.P.; Almeida, B.C.; Colwell, R.R.; Rivera, I.N. The importance of chitin in the marine environment. Mar. Biotechnol. 2011, 13, 823–830. [Google Scholar] [CrossRef]
- Clipson, N.; Otte, M.; Landy, E. Biogeochemical roles of fungi in marine and estuarine habitats. In Fungi in Biogeochemical Cycles; Gadd, Ed.; Cambridge University Press: Cambridge, UK, 2006; pp. 436–446. [Google Scholar]
- Barghini, P.; Moscatelli, D.; Garzillo, A.M.V.; Crognale, S.; Fenice, M. High production of cold-tolerant chitinases on shrimp wastes in bench-top bioreactor by the Antarctic fungus. Lecanicillium muscarium CCFEE 5003: Bioprocess optimization and characterization of two main enzymes. Enzyme Microb. Technol. 2013, 53, 331–338. [Google Scholar] [CrossRef]
- Felse, P.A.; Panda, T. Production of microbial chitinases. A revisit. Bioprocess. Eng. 2000, 23, 127–134. [Google Scholar] [CrossRef]
- Binod, P.; Sandhya, C.; Suma, P.; Szakacs, G.; Pandey, A. Fungal biosynthesis of endochitinase and chitobiase in solid state fermentation and their application for the production of N-acetyl-d-glucosamine from colloidal chitin. Bioresour. Technol. 2007, 98, 2742–2748. [Google Scholar] [CrossRef]
- Rocha-Pino, Z.; Vigueras, G.; Shirai, K. Production and activities of chitinases and hydrophobins from Lecanicillium lecanii. Bioproc. Biosyst. Eng. 2011, 34, 681–686. [Google Scholar] [CrossRef]
- Fenice, M.; Di Giambattista, R.; Leuba, J.L.; Federici, F. Inactivation of Mucor plumbeus by the combined action of chitinase and high hydrostatic pressure. Int. J. Food Microbiol. 1999, 52, 109–113. [Google Scholar] [CrossRef]
- Barghini, P.; Esti, M.; Pasqualetti, M.; Silvi, S.; Aquilanti, A.; Fenice, M. Crude cell wall degrading enzymes, by the Antarctic fungus Lecanicillium muscarium CCFEE 5003, inhibits the Ochratoxin-A producer Aspergillus carbonarius on white and red grapes. J. Environ. Prot. Ecol. 2013, 14, 1673–1679. [Google Scholar]
- Fenice, M.; Selbmann, L.; Di Giambattista, R.; Federici, F. Chitinolytic activity at low temperature of an Antarctic strain (A3) of Verticillium cfr. lecanii. Res. Microbiol. 1998, 149, 289–300. [Google Scholar] [CrossRef]
- Antal, Z.; Manczinger, L.; Szakacs, G.; Tengerdy, R.P.; Ferenczy, L. Colony growth, in vitro antagonism and secretion of extracellular enzymes in cold-tolerant strains of Trichoderma species. Mycol. Res. 2000, 104, 545–549. [Google Scholar] [CrossRef]
- Juarez-Jimenez, B.; Rodelas, B.; Martinez-Toledo, M.V.; Gonzalez-Lopez, J.; Crognale, S.; Gallo, A.M.; Pesciaroli, C.; Fenice, M. Production of chitinolytic enzymes by a strain (BM17) of Paenibacillus pabuli isolated from crab shells samples collected in the East Sector of Central Tyrrhenian Sea. Int. J. Biol. Macr. 2008, 43, 27–31. [Google Scholar] [CrossRef]
- Delgado-García, M.; Valdivia-Urdiales, B.; Aguilar-González, C.N.; Contreras-Esquivel, J.C.; Rodríguez-Herrera, R. Halophilic hydrolases as a new tool for the biotechnological industries. J. Sci. Food Agric. 2012, 92, 2575–2580. [Google Scholar] [CrossRef]
- Chung, D.; Baek, K.; Bae, S.S.; Jung, J. Identification and characterization of a marine-derived chitinolytic fungus, Acremonium sp. YS2-2. J. Microbiol. 2019, 57, 372–380. [Google Scholar] [CrossRef]
- Enache, M.; Kamekura, M. Hydrolytic enzymes of halophilic microorganisms and their economic values. Rom. J. Biochem. 2010, 47, 47–59. [Google Scholar]
- Yin, J.; Chen, J.C.; Wu, Q.; Chen, G.Q. (2015). Halophiles, coming stars for industrial biotechnology. Biotechnol. Adv. 2015, 33, 1433–1442. [Google Scholar] [CrossRef]
- Beygmoradi, A.; Homaei, A.; Hemmati, R.; Santos-Moriano, P.; Hormigo, D.; Fernández-Lucas, J. Marine chitinolytic enzymes, a biotechnological treasure hidden in the ocean? Appl. Microbiol. Biotechnol. 2018, 102, 9937–9948. [Google Scholar] [CrossRef]
- Domsch, K.H.; Gams, W.; Anderson, T. Compendium of Soil Fungi, 2nd ed.; IHW-Verlag: Eching, Germany, 2007. [Google Scholar]
- Chaverri, P.; Branco-Rocha, F.; Jaklitsch, W.; Gazis, R.; Degenkolb, T.; Samuels, G.J. Systematics of the Trichoderma harzianum species complex and the re-identification of commercial biocontrol strains. Mycologia 2015, 10, 558–590. [Google Scholar] [CrossRef]
- Saravanakumar, K.; Kathiresan, K.; Chen, J. Marine Fungal Genomics: Trichoderma. In Marine OMICS Principles and Applications; Kim, S.K., Ed.; CRC Press: London, UK, 2016; pp. 79–104. [Google Scholar]
- Touati, I.; Ruiz, N.; Thomas, O.; Druzhinina, I.S.; Atanasova, L.; Tabbene, O.; Elkahoui, S.; Benzekri, R.; Bouslama, L.; Pouchus, Y.F.; Limam, F. Hyporientalin A, an anti-Candida peptaibol from a marine Trichoderma orientale. World J. Microbiol. Biotechnol. 2018, 34, 98. [Google Scholar] [CrossRef]
- Mamarabadi, M.; Jensen, B.; Lübeck, M. Three endochitinase-encoding genes identified in the biocontrol fungus Clonostachys rosea are differentially expressed. Curr. Genet. 2008, 54, 57. [Google Scholar] [CrossRef]
- Zheng, Y.; Wang, X.; Liu, S.; Zhang, K.; Cai, Z.; Chen, X.; Zhang, Y.; Liu, J.; Wang, A. The Endochitinase of Clonostachys rosea Enhances the Biocontrol Efficiency of Bacillus amyloliquefaciens by Increasing Its Activities of Defense Enzymes. Int. J. Mol. Sci. 2018, 19, 2221. [Google Scholar]
- Dias, A.; Ruiz, N.; Couzinet-Mossion, A.; Bertrand, S.; Duflos, M.; Pouchus, Y.F.; Barnathan, G.; Nazih, H.; Wielgosz-Collin, G. The marine-derived fungus Clonostachys rosea, source of a rare conjugated 4-Me-6E, 8E-hexadecadienoic acid reducing viability of MCF-7 breast cancer cells and gene expression of lipogenic enzymes. Mar. Drugs 2015, 13, 4934–4948. [Google Scholar] [CrossRef]
- Fenice, M.; Barghini, P.; Selbmann, L.; Federici, F. Combined effects of agitation and aeration on the chitinolytic enzymes production by the Antarctic fungus Lecanicillium muscarium CCFEE 5003. Microb. Cell Fact. 2012, 11, 12. [Google Scholar] [CrossRef]
- Hull, S.R.; Yang, B.Y.; Venzke, D.; Kulhavy, K.; Montgomery, R. Composition of corn steep water during steeping. J. Agric. Food Chem. 1996, 44, 1857–1863. [Google Scholar] [CrossRef]
- Maddipati, P.; Atiyeh, H.K.; Bellmer, D.D.; Huhnke, R.L. Ethanol production from syngas by Clostridium strain P11 using corn steep liquor as a nutrient replacement to yeast extract. Bioresour. Technol. 2011, 102, 6494–6501. [Google Scholar] [CrossRef]
- Daniels, R.S. Corn Steep Liquor as A Biostimulant Composition. U.S. Patent No. 8568758 B2. Available online: http://www.freepatentsonline.com/y2012/0028801.html (accessed on 29 October 2013).
- De la Cruz, J.; Hidalgo-Gallego, A.; Lora, J.M.; Benitez, T.; Pintor-Toro, J.A.; Llobell, A. Isolation and characterization of three chitinases from Trichoderma harzianum. Eur. J. Biochem. 1992, 859–867. [Google Scholar] [CrossRef]
- Fenice, M.; Leuba, J.L.; Federici, F. Chitinolytic enzyme activity of Penicillium janthinellum P9 in bench-top bioreactor. J. Ferment. Bioeng. 1998, 86, 620–623. [Google Scholar] [CrossRef]
- Fenice, M.; Di Giambattista, R.; Raetz, E.; Leuba, J.L.; Federici, F. Repeated-batch and continuous production of chitinolytic enzymes by Penicillium janthinellum immobilised on chemically-modified macroporous cellulose. J. Biotechnol. 1998, 62, 119–131. [Google Scholar] [CrossRef]
- Kumar, M.; Brar, A.; Vivekanand, V.; Pareek, N. Production of chitinase from thermophilic Humicola grisea and its application in production of bioactive chitooligosaccharides. Int. J. Biol. Macromol. 2017, 104, 1641–1647. [Google Scholar] [CrossRef]
- Braga, F.R.; Soares, F.E.F.; Giuberti, T.Z.; Lopes, A.D.C.G.; Lacerda, T.; de Hollanda Ayupe, T.; Queiroz, P.V.; de Souza Gouveia, A.; Pinheiro, L.; Araujo, A.L.; et al. Nematocidal activity of extracellular enzymes produced by the nematophagous fungus Duddingtonia flagrans on cyathostomin infective larvae. Vet. Parasitol. 2015, 212, 214–218. [Google Scholar] [CrossRef]
- Panno, L.; Bruno, M.; Voyron, S.; Anastasi, A.; Gnavi, G.; Miserere, L.; Varese, G.C. Diversity, ecological role and potential biotechnological applications of marine fungi associated to the seagrass Posidonia Oceanicirca. New Biotechnol. 2013, 30, 685–694. [Google Scholar] [CrossRef]
- Ellis, M.B. Dematiaceous Hyphomycete; Commonwealth Mycological Institute: Kew, UK, 1971. [Google Scholar]
- Ellis, M.B. More Dematiaceous Hyphomycetes; Commonwealth Mycological Institute: Kew, UK, 1976. [Google Scholar]
- Kohlmeyer, J.; Kohlmeyer, E. Marine Mycology: The Higher Fungi; Academic Press: New York, NY, USA, 1979. [Google Scholar]
- Pitt, J.I. The genus Penicillium and its teleomorphic states Eupenicillium and Talaromyces; Academic Press Inc.: London, UK, 1979. [Google Scholar]
- Sutton, B.C. The Coelomycetes Fungi Imperfecti with Pycenidia, Acervuli and Stromata; Commonwealth Mycological Institute: Kew, UK, 1980. [Google Scholar]
- Klich, M.A. Identification of Common Aspergillus Species, 1st ed.; CBS: Utrecht, The Netherlands, 2002. [Google Scholar]
- White, T.J.; Bruns, T.; Lee, S.; Taylor, J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In PCR Protocols a Guide to Methods and Applications; Innis, M.A., Gelfand, D.H., Sninsky, J.J., White, T.J., Eds.; Academic Press: New York, NY, USA, 1990; pp. 315–322. [Google Scholar]
- Carbone, I.; Kohn, L.M. A method for designing primer sets for speciation studies in filamentous ascomycetes. Mycologia 1999, 91, 553–556. [Google Scholar] [CrossRef]
Sample Availability: Samples of the microbial strains are available from the authors. |



| Strain | Taxa | Substrate | Activity |
|---|---|---|---|
| IG132 | Alternaria chlamydospora | DD | − |
| IG135 | Arthrinium sp. | SC | − |
| IG133 | Aspergillus flavus | SC | +++ |
| IG105 | Aspergillus insuetus | PO | ng |
| IG136 | Aspergillus spelaeus | SC | ng |
| IG125 | Aspergillus versicolor | DD | − |
| IG118 | Cephalotrichum gorgonifer | PO | − |
| IG129 | Chaetomidium fimeti | DD | ++ |
| IG134 | Chaetomidium fimeti | SC | + |
| IG110 | Chaetomium sp. 1 | PO | ++ |
| IG107 | Chaetomium sp. 2 | PO | +++ |
| IG124 | Cladosporium sp. 1 | DD | − |
| IG101 | Cladosporium sp. 2 | PO | − |
| IG123 | Clonostachys rosea | PO | + |
| IG117 | Clonostachys rosea | PO | ++ |
| IG119 | Clonostachys rosea | PO | ++++ |
| IG120 | Fusarium sp. | PO | ng |
| IG100 | Mariannaea sp. | PO | ng |
| IG121 | Microascus brevicaulis | PO | ng |
| IG103 | Penicillium sp. | PO | − |
| IG113 | Pleospora sp. | PO | ng |
| IG122 | Scopulariopsis sp. | PO | − |
| IG126 | Stachybotrys chlorohalonatus | DD | +++ |
| IG127 | Trichoderma lixii | DD | ++++ |
| IG131 | Micelia sterilia 1 | DD | + |
| IG114 | Micelia sterilia 2 | PO | ng |
| IG108 | Micelia sterilia 3 | PO | ng |
| IG115 | Micelia sterilia 4 | PO | ng |
| Production (U/L) | Productivity (U/Lh) | |||||
| Coefficient | RC | SE | P | RC | SE | P |
| Constant | 3.5158 | 0.1112 | 1.10 × 10−13 | 3.4515 | 0.1224 | 4.7937 × 10−13 |
| CSL | 0.8666 | 0.0688 | 1.17 × 10−8 | 0.9656 | 0.0758 | 1.0124 × 10−8 |
| YNB | 0.6908 | 0.0688 | 1.72 × 10−7 | 0.5917 | 0.0758 | 2.9116 × 10−6 |
| CSL*CSL | −0.4204 | 0.1139 | 2.71 × 10−3 | −0.1911 | 0.1254 | 1.5150 × 10−1 |
| YNB*YNB | −0.5110 | 0.1139 | 6.13 × 10−4 | −0.5601 | 0.1254 | 6.3504 × 10−4 |
| CSL*YNB | −0.2251 | 0.0843 | 1.92 × 10−2 | −0.0978 | 0.0928 | 3.1117 × 10−1 |
| ANOVA table (Modde 5) | ||||||
| F value | 60.7442 | P = 0.000 | 49.656 | P = 0.000 | ||
| Q2 | 0.92 | 0.895 | ||||
| R2 | 0.959 | 0.95 | ||||
| R2 adjusted | 0.943 | 0.931 | ||||
| Experiment | CSL (%) | YNB (%) |
|---|---|---|
| N1, N12 | 0 | 0 |
| N2, N13 | 0.5 | 0 |
| N3, N14 | 0 | 0.5 |
| N4, N15 | 0.5 | 0.5 |
| N5, N16 | 0 | 0.25 |
| N6, N17 | 0.5 | 0.25 |
| N7, N18 | 0.25 | 0 |
| N8, N19 | 0.25 | 0.5 |
| N9, N10, N11 | 0.25 | 0.25 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pasqualetti, M.; Barghini, P.; Giovannini, V.; Fenice, M. High Production of Chitinolytic Activity in Halophilic Conditions by a New Marine Strain of Clonostachys rosea. Molecules 2019, 24, 1880. https://doi.org/10.3390/molecules24101880
Pasqualetti M, Barghini P, Giovannini V, Fenice M. High Production of Chitinolytic Activity in Halophilic Conditions by a New Marine Strain of Clonostachys rosea. Molecules. 2019; 24(10):1880. https://doi.org/10.3390/molecules24101880
Chicago/Turabian StylePasqualetti, Marcella, Paolo Barghini, Valeria Giovannini, and Massimiliano Fenice. 2019. "High Production of Chitinolytic Activity in Halophilic Conditions by a New Marine Strain of Clonostachys rosea" Molecules 24, no. 10: 1880. https://doi.org/10.3390/molecules24101880
APA StylePasqualetti, M., Barghini, P., Giovannini, V., & Fenice, M. (2019). High Production of Chitinolytic Activity in Halophilic Conditions by a New Marine Strain of Clonostachys rosea. Molecules, 24(10), 1880. https://doi.org/10.3390/molecules24101880