Fluorescent DNA Biosensor for Single-Base Mismatch Detection Assisted by Cationic Comb-Type Copolymer
Abstract
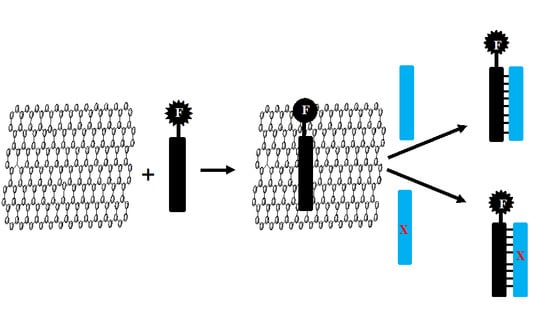
:1. Introduction
2. Materials and Methods
2.1. Reagents and Materials
2.2. Fluorescence Spectroscopy
2.3. UV-Melting Point (Tm) Measurement
2.4. Cell Culture
3. Results and Discussion
4. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Kashuk, C.; SenGupta, S.; Eichler, E.; Chakravarti, A. ViewGene: A graphical tool for polymorphism visualization and characterization. Genome Res. 2002, 12, 333–338. [Google Scholar] [CrossRef] [PubMed]
- Brennan, M.D. High throughput genotyping technologies for pharmacogenomics. Am. J. Pharm. 2001, 1, 295–302. [Google Scholar] [CrossRef]
- Kwok, P.Y. Methods for genotyping single nucleotide polymorphisms. Annu. Rev. Genom. Hum. Genet. 2001, 2, 235–258. [Google Scholar] [CrossRef] [PubMed]
- Chicurel, M. Faster, better, cheaper genotyping. Nature 2001, 412, 580–582. [Google Scholar] [CrossRef] [PubMed]
- Kim, W.J.; Sato, Y.; Akaike, T.; Maruyama, A. Cationic comb-type copolymers for DNA analysis. Nat. Mater. 2003, 2, 815–820. [Google Scholar] [CrossRef] [PubMed]
- Kim, W.J.; Akaike, T.; Maruyama, A. DNA strand exchange stimulated by spontaneous complex formation with cationic comb-type copolymer. J. Am. Chem. Soc. 2002, 124, 12676–12677. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Luan, G.; Guo, Q.; Liang, J. A new class of homogeneous nucleic acid probes based on specific displacement hybridization. Nucleic Acids Res. 2002, 30, e5. [Google Scholar] [CrossRef]
- Lu, C.H.; Yang, H.H.; Zhu, C.L.; Chen, X.; Chen, G.N. A graphene platform for sensing biomolecules. Angew. Chem. Int. Ed. 2009, 121, 4879–4881. [Google Scholar] [CrossRef]
- Jang, H.J.; Kim, Y.K.; Kwon, H.M.; Yeo, W.S.; Kim, D.E.; Min, D.H. A Graphene-Based Platform for the Assay of Duplex-DNA Unwinding by Helicase. Angew. Chem. Int. Ed. 2010, 122, 5839–5843. [Google Scholar] [CrossRef]
- Wang, Y.; Li, Z.H.; Hu, D.H.; Lin, C.T.; Li, J.H.; Lin, Y.H. Aptamer/Graphene Oxide Nanocomplex for in Situ Molecular Probing in Living Cells. J. Am. Chem. Soc. 2010, 132, 9274–9276. [Google Scholar] [CrossRef]
- He, S.J.; Song, B.; Li, D.; Zhu, C.F.; Qi, W.P.; Wen, Y.Q.; Wang, L.H.; Song, S.P.; Fang, H.P.; Fan, C.H. A graphene nanoprobe for rapid, sensitive, and multicolor fluorescent DNA analysis. Adv. Funct. Mater. 2010, 20, 453–459. [Google Scholar] [CrossRef]
- Song, Y.J.; Qu, K.G.; Zhao, C.; Ren, J.S.; Qu, X.G. Graphene Oxide: Intrinsic Peroxidase Catalytic Activity and Its Application to Glucose Detection. Adv. Mater. 2010, 22, 2206–2210. [Google Scholar] [CrossRef] [PubMed]
- Huang, R.C.; Chiu, W.J.; Li, Y.J.; Huang, C.C. Detection of microRNA in Tumor Cells using Exonuclease III and Graphene Oxide-Regulated Signal Amplification. ACS Appl. Mater. Interfaces 2014, 6, 21780–21787. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Yang, H.Y.; Ai, Y. DNA single-base mismatch study using graphene oxide nanosheets-based fluorometric biosensors. Anal. Chem. 2015, 87, 9132–9136. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Huang, Y.; Wang, D.; Song, B.; Li, Z.; Song, S.; Wang, L.; Jiang, B.; Zhao, X.; Yan, J.; et al. A power-free microfluidic chip for SNP genotyping using graphene oxide and a DNA intercalating dye. Chem. Commun. 2013, 49, 3125–3127. [Google Scholar] [CrossRef]
- Wang, H.; Chen, T.; Wu, S.; Chu, X.; Yu, R. A novel biosensing strategy for screening G-quadruplex ligands based on graphene oxide sheets. Biosens. Bioelectron. 2012, 34, 88–93. [Google Scholar] [CrossRef] [PubMed]
- Wen, Y.Q.; Xing, F.F.; He, S.J.; Song, S.P.; Wang, L.H.; Long, Y.T.; Li, D.; Fan, C.H. A graphene-based fluorescent nanoprobe for silver(I) ions detection by using graphene oxide and a silver-specific oligonucleotide. Chem. Commun. 2010, 46, 2596–2598. [Google Scholar] [CrossRef]
- Chang, H.X.; Tang, L.H.; Wang, Y.; Jang, J.H.; Li, J.H. Graphene Fluorescence Resonance Energy Transfer Aptasensor for the Thrombin Detection. Anal. Chem. 2010, 82, 2341–2346. [Google Scholar] [CrossRef]
- Park, J.W.; Tatavarty, R.; Kim, D.W.; Jung, H.T.; Gu, M.B. Immobilization-free screening of aptamers assisted by graphene oxide. Chem. Commun. 2012, 48, 2071–2073. [Google Scholar] [CrossRef]
- Zhang, L.M.; Xia, J.G.; Zhao, Q.H.; Liu, L.W.; Zhang, Z.J. Functional graphene oxide as a nanocarrier for controlled loading and targeted delivery of mixed anticancer drugs. Small 2010, 6, 537–544. [Google Scholar] [CrossRef]
- Peng, C.; Hu, W.B.; Zhou, Y.T.; Fan, C.H. Intracellular Imaging with a Graphene-Based Fluorescent Probe. Small 2010, 6, 1686–1692. [Google Scholar] [CrossRef] [PubMed]
- Qu, K.; Fang, M.; Zhang, S.; Liu, H.; Zeng, X. A redox conjugated polymer-based all-solid-state reference electrode. Polymers 2018, 10, 1191. [Google Scholar] [CrossRef]
- Lee, D.; Sang, J.; Yoo, P.; Shin, T.; Oh, K.; Park, J. Machine-Washable Smart Textiles with Photothermal and Antibacterial Activities from Nanocomposite Fibers of Conjugated Polymer Nanoparticles and Polyacrylonitrile. Polymers 2019, 11, 16. [Google Scholar] [CrossRef]
- Liu, F.; Zhang, Y.; Wang, H.; Zhang, S. Novel Conjugated Polymers Prepared by Direct (Hetero) arylation: An Eco-Friendly Tool for Organic Electronics. Molecules 2018, 23, 408. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Yu, F.; Zhang, Z.; Chen, Y.; Du, J. Highly sensitive self-complementary DNA nanoswitches triggered by polyelectrolytes. Nanoscale 2016, 8, 464–470. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Wu, Y.; Yu, F.; Niu, C.; Du, Z.; Chen, Y.; Du, J. Rapid and annealing-free self-assembly of DNA building blocks for 3D hydrogel chaperoned by cationic comb-type copolymers. J. Biomater. Sci. Polym. Ed. 2017, 28, 1511–1524. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Han, J.L.; Pei, Y.X.; Fan, R.Z.; Du, J. Chaperone copolymer assisted aptamer-patterned DNA hydrogels for triggering spatiotemporal release of protein. ACS Appl. Bio Mater. 2018, in press. [Google Scholar] [CrossRef]
- Maruyama, A.; Katoh, M.; Ishihara, T.; Akaike, T. Comb-type polycations effectively stabilize DNA triplex. Bioconjug. Chem. 1997, 8, 3–6. [Google Scholar] [CrossRef]
Sample Availability: Samples of the compounds PLL-g-Dex and GO are available from the authors. |






| Oligonucleotide | Sequence |
|---|---|
| T-DNA | 5′-TAMRA-TCTTTGGGACCACTGTCG-3′ |
| cDNA | 5′-CGACAGTGGTCCCAAAGA-3′ |
| M1 (C-T) | 5′-CGACAGTGTTCCCAAAGA-3′ |
| M2 (C-A) | 5′-CGACAGTGATCCCAAAGA-3′ |
| M3 (C-C) | 5′-CGACAGTGCTCCCAAAGA-3′ |
| Double-Strands DNA | Tm (without PLL-g-Dex) | Tm (with PLL-g-Dex) | ||||
|---|---|---|---|---|---|---|
| Heating | Cooling | Average | Heating | Cooling | Average | |
| T-DNA/cDNA | 54.68 | 57.34 | 56.01 | 64.15 | 66.37 | 65.26 |
| T-DNA/M1 | 46.85 | 48.41 | 47.63 | 52.68 | 54.74 | 53.71 |
| T-DNA/M2 | 47.68 | 49.48 | 48.58 | 54.86 | 55.60 | 55.23 |
| T-DNA/M3 | 49.45 | 50.01 | 49.74 | 49.65 | 50.39 | 50.02 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Han, J.; Wu, J.; Du, J. Fluorescent DNA Biosensor for Single-Base Mismatch Detection Assisted by Cationic Comb-Type Copolymer. Molecules 2019, 24, 575. https://doi.org/10.3390/molecules24030575
Han J, Wu J, Du J. Fluorescent DNA Biosensor for Single-Base Mismatch Detection Assisted by Cationic Comb-Type Copolymer. Molecules. 2019; 24(3):575. https://doi.org/10.3390/molecules24030575
Chicago/Turabian StyleHan, Jialun, Jincai Wu, and Jie Du. 2019. "Fluorescent DNA Biosensor for Single-Base Mismatch Detection Assisted by Cationic Comb-Type Copolymer" Molecules 24, no. 3: 575. https://doi.org/10.3390/molecules24030575
APA StyleHan, J., Wu, J., & Du, J. (2019). Fluorescent DNA Biosensor for Single-Base Mismatch Detection Assisted by Cationic Comb-Type Copolymer. Molecules, 24(3), 575. https://doi.org/10.3390/molecules24030575