Ultrashort Cationic Lipopeptides–Effect of N-Terminal Amino Acid and Fatty Acid Type on Antimicrobial Activity and Hemolysis
Abstract
:1. Introduction
2. Results and Discussion
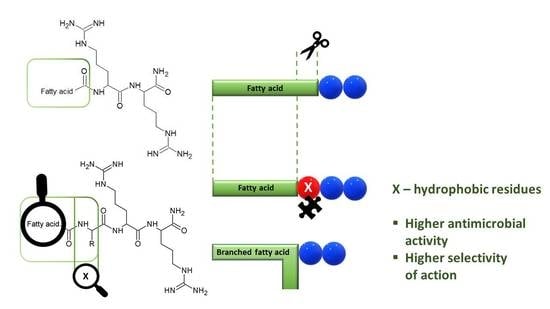
2.1. Selection of a Position to be Substituted
2.2. Hydrophobicity and Antimicrobial Activity of Reference Lipopeptides
2.3. Hydrophobicity of Lipopeptide Series
2.4. Peptide Hydrophobicity vs. Antimicrobial Activity and Hemolysis
2.5. Antimicrobial Activity of Selected Lipopeptides against Reference ESKAPE Strains
3. Materials and Methods
3.1. Peptide Synthesis
3.2. Determination of Peptide Hydrophobicity with RP-HPLC
3.3. Antimicrobial Activity
3.3.1. Cultivation of Microorganisms
3.3.2. Activity against Planktonic Cultures
3.4. Hemolysis Assay
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| ACN | acetonitrile |
| Acm | S-acetamidomethyl group |
| ATCC | American Type Culture Collection |
| Boc | tert-butyloxycarbonyl group |
| CLSI | Clinical and Laboratory Standards Institute |
| DCM | dichloromethane |
| DIC | N,N′-diisopropylcarbodiimide |
| DMF | N,N-dimethylformamide |
| EDT | 1,2-ethanedithiol |
| EDTA | ethylenediaminetetraacetic acid |
| ESI–MS | electrospray-ionization mass spectrometry |
| Fmoc | 9-fluorenylmethoxycarbonyl group |
| HC50 | lipopeptide concentration causing 50% hemolysis |
| hRBCs | human red blood cells |
| Met(O)/M(O) | methionine sulfoxide |
| Met(O2)/M(O2) | methionine sulfone |
| MIC | minimum inhibitory concentration |
| MOPS | 3-(N-morpholino)propanesulfonic acid |
| Nle | norleucine |
| Nva | norvaline |
| Pbf | 2,2,4,6,7-pentamethyl-dihydrobenzofuran-5-sulfonyl residue |
| PBS | phosphate-buffer saline |
| RP-HPLC | reverse-phase high-performance liquid chromatography |
| tBu | tert-butyl group |
| TFA | trifluoroacetic acid |
| TIS | triisopropylsilane |
| Trt | trityl group |
| USCLs | ultrashort cationic lipopeptides |
References
- Mnif, I.S.; Ghribi, D. Review Lipopeptides Biosurfactants: Mean Classes and New Insights for Industrial, Biomedical, and Environmental Applications. Inc. Biopolymers. (Pept Sci.) 2015, 104, 129–147. [Google Scholar] [CrossRef] [PubMed]
- Makovitzki, A.; Avrahami, D.; Shai, Y.; Sela, M. Ultrashort antibacterial and antifungal lipopeptides. Proc. Natl. Acad. Sci. USA 2006, 103, 15997–16002. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Greber, K.E.; Dawgul, M.; Kamysz, W.; Sawicki, W. Cationic Net Charge and Counter Ion Type as Antimicrobial Activity Determinant Factors of Short Lipopeptides. Front. Microbiol. 2017, 8, 123. [Google Scholar] [CrossRef] [Green Version]
- Maciejewska, M.; Bauer, M.; Neubauer, D.; Kamysz, W.; Dawgul, M. Influence of Amphibian Antimicrobial Peptides and Short Lipopeptides on Bacterial Biofilms Formed on Contact Lenses. Materials 2016, 9, 873. [Google Scholar] [CrossRef] [Green Version]
- Dawgul, M.; Greber, K.; Bartoszewska, S.; Baranska-Rybak, W.; Sawicki, W.; Kamysz, W. In Vitro Evaluation of Cytotoxicity and Permeation Study on Lysine- and Arginine-Based Lipopeptides with Proven Antimicrobial Activity. Molecules 2017, 22, 2173. [Google Scholar] [CrossRef] [Green Version]
- Alves, D.; Magalhães, A.; Grzywacz, D.; Neubauer, D.; Kamysz, W.; Pereira, M.O. Co-immobilization of Palm and DNase I for the development of an effective anti-infective coating for catheter surfaces. Acta Biomater. 2016, 44, 313–322. [Google Scholar] [CrossRef] [Green Version]
- Pendleton, J.N.; Gorman, S.P.; Gilmore, B.F. Clinical relevance of the ESKAPE pathogens. Expert Rev. Anti. Infect. Ther. 2013, 11, 297–308. [Google Scholar] [CrossRef]
- Mulani, M.S.; Kamble, E.E.; Kumkar, S.N.; Tawre, M.S.; Pardesi, K.R. Emerging Strategies to Combat ESKAPE Pathogens in the Era of Antimicrobial Resistance: A Review. Front. Microbiol. 2019, 10, 539. [Google Scholar] [CrossRef]
- Armas, F.; Pacor, S.; Ferrari, E.; Guida, F.; Pertinhez, T.A.; Romani, A.A.; Scocchi, M.; Benincasa, M. Design, antimicrobial activity and mechanism of action of Arg-rich ultra-short cationic lipopeptides. PLoS ONE 2019, 14, e0212447. [Google Scholar] [CrossRef]
- Mangoni, M.L.; Shai, Y. Short native antimicrobial peptides and engineered ultrashort lipopeptides: Similarities and differences in cell specificities and modes of action. Cell. Mol. Life Sci. 2011, 68, 2267–2280. [Google Scholar] [CrossRef]
- Sader, H.S.; Farrell, D.J.; Flamm, R.K.; Jones, R.N. Daptomycin activity tested against 164457 bacterial isolates from hospitalised patients: Summary of 8 years of a Worldwide Surveillance Programme (2005–2012). Int. J. Antimicrob. Agents 2014, 43, 465–469. [Google Scholar] [CrossRef] [PubMed]
- Arendrup, M.C.; Perlin, D.S. Echinocandin resistance: An emerging clinical problem? Curr. Opin. Infect. Dis. 2014, 27, 484–492. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Błażewicz, I.; Jaśkiewicz, M.; Piechowicz, L.; Neubauer, D.; Nowicki, R.J.; Kamysz, W.; Barańska-Rybak, W. Increasing rate of daptomycin non-susceptible strains of Staphylococcus aureus in patients with atopic dermatitis. Adv. Dermatol. Allergol. 2017, 34, 547–552. [Google Scholar] [CrossRef] [PubMed]
- Gales, A.C.; Jones, R.N.; Sader, H.S. Global assessment of the antimicrobial activity of polymyxin B against 54 731 clinical isolates of Gram-negative bacilli: Report from the SENTRY antimicrobial surveillance programme (2001–2004). Clin. Microbiol. Infect. 2006, 12, 315–321. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Straus, S.K.; Hancock, R.E.W. Mode of action of the new antibiotic for Gram-positive pathogens daptomycin: Comparison with cationic antimicrobial peptides and lipopeptides. Biochim. Biophys. Acta 2006, 1758, 1215–1223. [Google Scholar] [CrossRef] [Green Version]
- Laverty, G.; McLaughlin, M.; Shaw, C.; Gorman, S.P.; Gilmore, B.F. Antimicrobial Activity of Short, Synthetic Cationic Lipopeptides. Chem. Biol. Drug Des. 2010, 75, 563–569. [Google Scholar] [CrossRef]
- Dawgul, M.; Baranska-Rybak, W.; Kamysz, E.; Karafova, A.; Nowicki, R.; Kamysz, W. Activity of short lipopeptides and conventional antimicrobials against planktonic cells and biofilms formed by clinical strains of Staphylococcus aureus. Future Med. Chem. 2012, 4, 1541–1551. [Google Scholar] [CrossRef]
- Dawgul, M.; Maciejewska, M.; Jaskiewicz, M.; Karafova, A.; Kamysz, W. Antimicrobial peptides as potential tool to fight bacterial biofilm. Acta Pol. Pharm. 2014, 71, 39–47. [Google Scholar]
- Li, L.; Vorobyov, I.; Allen, T.W. The different interactions of lysine and arginine side chains with lipid membranes. J. Phys. Chem. B 2013, 117, 11906–11920. [Google Scholar] [CrossRef]
- Chen, Y.; Guarnieri, M.T.; Vasil, A.I.; Vasil, M.L.; Mant, C.T.; Hodges, R.S. Role of peptide hydrophobicity in the mechanism of action of α-helical antimicrobial peptides. Antimicrob. Agents Chemother. 2007, 51, 1398–1406. [Google Scholar] [CrossRef] [Green Version]
- Malina, A.; Shai, Y. Conjugation of fatty acids with different lengths modulates the antibacterial and antifungal activity of a cationic biologically inactive peptide. Biochem. J. 2005, 390, 695–702. [Google Scholar] [CrossRef] [PubMed]
- Hamley, I.W. Lipopeptides: From self-assembly to bioactivity. Chem. Commun. 2015, 51, 8574–8583. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zavascki, A.P.; Goldani, L.Z.; Li, J.; Nation, R.L. Polymyxin B for the treatment of multidrug-resistant pathogens: A critical review. J. Antimicrob. Chemother. 2007, 60, 1206–1215. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tripet, B.; Cepeniene, D.; Kovacs, J.M.; Mant, C.T.; Krokhin, O.V.; Hodges, R.S. Requirements for prediction of peptide retention time in reversed-phase high-performance liquid chromatography: Hydrophilicity/hydrophobicity of side-chains at the N- and C-termini of peptides are dramatically affected by the end-groups and location. J. Chromatogr. A 2007, 1141, 212–225. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Domalaon, R.; Yang, X.; O’Neil, J.; Zhanel, G.G.; Mookherjee, N.; Schweizer, F. Structure-activity relationships in ultrashort cationic lipopeptides: The effects of amino acid ring constraint on antibacterial activity. Amino Acids 2014, 46, 2517–2530. [Google Scholar] [CrossRef]
- Nasompag, S.; Dechsiri, P.; Hongsing, N.; Phonimdaeng, P.; Daduang, S.; Klaynongsruang, S.; Camesano, T.A.; Patramanon, R. Effect of acyl chain length on therapeutic activity and mode of action of the CX-KYR-NH2 antimicrobial lipopeptide. Biochim. Biophys. Acta 2015, 1848, 2351–2364. [Google Scholar] [CrossRef] [Green Version]
- Sereda, T.J.; Mant, C.T.; Quinn, A.M.; Hodges, R.S. Effect of the α-amino group on peptide retention behaviour in reversed-phase chromatography Determination of the pKa values of the α-amino group of 19 different N-terminal amino acid residues. J. Chromatogr. 1993, 646, 17–30. [Google Scholar] [CrossRef]
- Lao, Y.W.; Gungormusler-Yilmaz, M.; Shuvo, S.; Verbeke, T.; Spicer, V.; Krokhin, O.V. Chromatographic behavior of peptides containing oxidized methionine residues in proteomic LC-MS experiments: Complex tale of a simple modification. J. Proteomics 2015, 125, 131–139. [Google Scholar] [CrossRef] [PubMed]
- Meyer, V.R. Reversed-Phase Chromatography. In Practical High-Performance Liquid Chromatography; John Wiley & Sons, Ltd.: Chichester, UK, 2010; pp. 173–193. [Google Scholar]
- Cruz, E.; Euerby, M.R.; Johnson, C.M.; Hackett, C.A. Chromatographic classification of commercially available reverse-phase HPLC columns. Chromatographia 1997, 44, 151–161. [Google Scholar] [CrossRef]
- Kimata, K.; Iwaguchi, K.; Onishi, S.; Jinno, K.; Eksteen, R.; Hosoya, K.; Araki, M.; Tanaka, N. Chromatographic characterization of silica c18 packing materials. correlation between a preparation method and retention behavior of stationary phase. J. Chromatogr. Sci. 1989, 27, 721–728. [Google Scholar] [CrossRef]
- Very Simple IC50 Tool Kit—Calculate, Measure, Determine IC50 Online. Available online: http://www.ic50.tk/index.html (accessed on 14 October 2019).
- Greber, K.E.; Ciura, K.; Belka, M.; Kawczak, P.; Nowakowska, J.; Bączek, T.; Sawicki, W. Characterization of antimicrobial and hemolytic properties of short synthetic cationic lipopeptides based on QSAR/QSTR approach. Amino Acids 2018, 50, 479–485. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- D.R. Lide Handbook of Chemistry and Physics, 72nd ed.; CRC Press: Boca Raton, FL, USA, 1991. [Google Scholar]
- Lohan, S.; Cameotra, S.S.; Bisht, G.S. Systematic study of non-natural short cationic lipopeptides as novel broad-spectrum antimicrobial agents. Chem. Biol. Drug Des. 2013, 82, 557–566. [Google Scholar] [CrossRef] [PubMed]
- Liao, S.M.; Du, Q.S.; Meng, J.Z.; Pang, Z.W.; Huang, R.B. The multiple roles of histidine in protein interactions. Chem. Cent. J. 2013, 7, 44. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Heyda, J.; Mason, P.E.; Jungwirth, P. Attractive Interactions between Side Chains of Histidine-Histidine and Histidine-Arginine-Based Cationic Dipeptides in Water. J. Phys. Chem. B 2010, 114, 8744–8749. [Google Scholar] [CrossRef]
- Zhou, P.; Tian, F.; Lv, F.; Shang, Z. Geometric characteristics of hydrogen bonds involving sulfur atoms in proteins. Proteins. 2009, 76, 151–163. [Google Scholar] [CrossRef]
- Jaśkiewicz, M.; Neubauer, D.; Kamysz, W. Comparative Study on Antistaphylococcal Activity of Lipopeptides in Various Culture Media. Antibiotics 2017, 6, 15. [Google Scholar] [CrossRef] [Green Version]
- Snyder, D.S.; McIntosh, T.J. The lipopolysaccharide barrier: Correlation of antibiotic susceptibility with antibiotic permeability and fluorescent probe binding kinetics. Biochemistry 2000, 39, 11777–11787. [Google Scholar] [CrossRef]
- Nikaido, H.; Vaara, M. Molecular basis of bacterial outer membrane permeability. Microbiol. Rev. 1985, 49, 1–32. [Google Scholar] [CrossRef] [Green Version]
- Gruenheid, S.; Le Moual, H. Resistance to antimicrobial peptides in Gram-negative bacteria. FEMS Micrbiol. Lett. 2012, 330, 81–89. [Google Scholar] [CrossRef] [Green Version]
- Gunn, J.S.; Lim, K.B.; Krueger, J.; Kim, K.; Guo, L.; Hackett, M.; Miller, S.I. PmrA-PmrB-regulated genes necessary for 4-aminoarabinose lipid A modification and polymyxin resistance. Mol. Microbiol. 1998, 27, 1171–1182. [Google Scholar] [CrossRef]
- Padilla, E.; Llobet, E.; Doménech-Sánchez, A.; Martínez-Martínez, L.; Bengoechea, J.A.; Albertí, S. Klebsiella pneumoniae AcrAB efflux pump contributes to antimicrobial resistance and virulence. Antimicrob. Agents Chemother. 2010, 54, 177–183. [Google Scholar] [CrossRef] [Green Version]
- Marcoleta, A.E.; Varas, M.A.; Ortiz-Severín, J.; Vásquez, L.; Berríos-Pastén, C.; Sabag, A.V.; Chávez, F.P.; Allende, M.L.; Santiviago, C.A.; Monasterio, O.; et al. Evaluating different virulence traits of Klebsiella pneumoniae using Dictyostelium discoideum and zebrafish larvae as host models. Front. Cell. Infect. Microbiol. 2018, 8, 30. [Google Scholar] [CrossRef]
- Formosa, C.; Herold, M.; Vidaillac, C.; Duval, R.E.; Dague, E. Unravelling of a mechanism of resistance to colistin in Klebsiella pneumoniae using atomic force microscopy. J. Antimicrob. Chemother. 2015, 70, 2261–2270. [Google Scholar] [CrossRef] [Green Version]
- Campos, M.A.; Vargas, M.A.; Regueiro, V.; Llompart, C.M.; Albertí, S.; Bengoechea, J.A. Capsule polysaccharide mediates bacterial resistance to antimicrobial peptides. Infect. Immun. 2004, 72, 7107–7114. [Google Scholar] [CrossRef] [Green Version]
- Dé, E.; Baslé, A.; Jaquinod, M.; Saint, N.; Malléa, M.; Molle, G.; Pagès, J.M. A new mechanism of antibiotic resistance in Enterobacteriaceae induced by a structural modification of the major porin. Mol. Microbiol. 2001, 41, 189–198. [Google Scholar] [CrossRef]
- Gayet, S.; Chollet, R.; Molle, G.; Pagès, J.M.; Chevalier, J. Modification of outer membrane protein profile and evidence suggesting an active drug pump in Enterobacter aerogenes clinical strains. Antimicrob. Agents Chemother. 2003, 47, 1555–1559. [Google Scholar] [CrossRef] [Green Version]
- Joshi, S.; Mumtaz, S.; Singh, J.; Pasha, S.; Mukhopadhyay, K. Novel Miniature Membrane Active Lipopeptidomimetics against Planktonic and Biofilm Embedded Methicillin-Resistant Staphylococcus aureus. Sci. Rep. 2018, 8, 1021. [Google Scholar] [CrossRef] [Green Version]
- Clinical and Laboratory Standards Institute (CLSI). Reference Method for Broth Dilution Antifungal Susceptibility Testing of Yeasts; Approved Standards-Second Edition; CLSI Document M27-2A 2002; CLSI: Wayne, PA, USA, 2002; Volume 22, ISBN 1562384694. [Google Scholar]
- Clinical and Laboratory Standards Institute (CLSI). Methods for Dilution Antimicrobial Susceptibility Tests for Bacteria That Grow Aerobically; Approved Standard—Ninth Edition; CLSI: Wayne, PA, USA, 2012; Volume 32, ISBN 1562387839. [Google Scholar]
- Sikora, K.; Jaśkiewicz, M.; Neubauer, D.; Bauer, M.; Bartoszewska, S.; Barańska-Rybak, W.; Kamysz, W. Counter-ion effect on antistaphylococcal activity and cytotoxicity of selected antimicrobial peptides. Amino Acids 2018, 50, 609–619. [Google Scholar] [CrossRef] [Green Version]
- Avrahami, D.; Shai, Y. A New Group of Antifungal and Antibacterial Lipopeptides Derived from Non-membrane Active Peptides Conjugated to Palmitic Acid. J. Biol. Chem. 2004, 279, 12277–12285. [Google Scholar] [CrossRef] [Green Version]
- Reinhardt, A.; Neundorf, I. Design and Application of Antimicrobial Peptide Conjugates. Int. J. Mol. Sci. 2016, 17, 701. [Google Scholar] [CrossRef]
- Albada, H.B.; Prochnow, P.; Bobersky, S.; Langklotz, S.; Schriek, P.; Bandow, J.E.; Metzler-Nolte, N. Tuning the activity of a short arg-trp antimicrobial peptide by lipidation of a C- or N-terminal lysine side-chain. ACS Med. Chem. Lett. 2012, 3, 980–984. [Google Scholar] [CrossRef] [Green Version]
- Wenda, J.M.; Juhaniewicz, J.; Tymecka, D.; Konarzewska, D.; Sȩk, S. Modulation of Activity of Ultrashort Lipopeptides toward Negatively Charged Model Lipid Films. Langmuir 2017, 33, 4619–4627. [Google Scholar] [CrossRef]
- Papo, N.; Oren, Z.; Pag, U.; Sahl, H.-G.; Shai, Y. The consequence of sequence alteration of an amphipathic alpha-helical antimicrobial peptide and its diastereomers. J. Biol. Chem. 2002, 277, 33913–33921. [Google Scholar] [CrossRef] [Green Version]
Sample Availability: Samples of the compounds are available from the authors. |





| Code | Sequence | tR’ [min] | ∆tR [min] * |
|---|---|---|---|
| 24 | C10-FRR-NH2 | 34.68 | 10.38 |
| 106 | C10-RFR-NH2 | 31.44 | 7.14 |
| 108 | C10-RRF-NH2 | 31.07 | 6.77 |
| 25 | C12-FRR-NH2 | 41.45 | 8.87 |
| 107 | C12-RFR-NH2 | 38.67 | 6.09 |
| 109 | C12-RRF-NH2 | 38.02 | 5.44 |
| Ile | Leu | Nle | |||
|---|---|---|---|---|---|
| Structure of the Side Chain | -CH2-CH(CH3)CH2CH3 | -CH2CH2CH(CH3)CH3 | -CH2CH2CH2CH2CH3 | ||
| nC | Adjusted retention time [min] | ||||
| 8 | 24.017 | < | 25.032 | < | 25.429 |
| 10 | 31.953 | 32.846 | 33.155 | ||
| 12 | 39.047 | 39.821 | 39.958 | ||
| 14 | 45.650 | 46.441 | 46.681 | ||
| Code | Name | X | tR’ [min] | HC50 [µg/mL] | MIC SA [µg/mL] (SI) | MIC PA [µg/mL] (SI) | MIC CA [µg/mL] (SI) |
|---|---|---|---|---|---|---|---|
| 1 | C8-RR-NH2 | Reference lipopeptides | 14.46 | - | >256 | >256 | >256 |
| 2 | C10-RR-NH2 | 24.30 | - | >256 | >256 | >256 | |
| 3 | C12-RR-NH2 | 32.58 | >256 | 32 (>8) | 128 (>2) | 64 (>4) | |
| 4 | C14-RR-NH2 | 39.79 | 68.43 (±1.41) | 8 * (8.55) * | 32 (2.14) | 8 (8.55) | |
| 5 | C16-RR-NH2 | 46.51 | 22.91 (±1.18) | 8 (2.86) | 16 * (1.43) * | 4 (5.73) | |
| 6 | C18-RR-NH2 | 53.39 | 25.19 (±0.67) | 16 (1.57) | 64 (0.39) | 4 * (6.30) * | |
| 10 | C14-ARR-NH2 | A | 40.92 | 41.08 (±0.62) | 8 (5.14) | 32 (1.28) | 16 (2.57) |
| 14 | C14-C(Acm)RR-NH2 | C(Acm) | 43.24 | 39.72 (±1.25) | 16 (2.48) | 16 (2.48) | 8 (4.97) |
| 18 | C14-DRR-NH2 | D | 40.25 | 49.95 (±1.00) | 64 (0.78) | 128 (0.46) | 32 (1.86) |
| 22 | C14-ERR-NH2 | E | 40.17 | 59.51 (±4.00) | 64 (0.93) | 128 (0.46) | 32 (1.86) |
| 24 | C10-FRR-NH2 | F | 34.68 | >256 | 16 (>16) | 32 (>8) | 64 (>4) |
| 25 | C12-FRR-NH2 | F | 41.45 | 91.21 (±16.38) | 4 (22.80) | 8 (11.40) | 8 (11.40) |
| 26 | C14-FRR-NH2 | F | 47.75 | 39.09 (±1.18) | 4 (9.77) | 8 (4.89) | 2 (19.55) |
| 29 | C12-GRR-NH2 | G | 33.20 | >256 | 32 (>8) | 128 (>2) | 64 (>4) |
| 30 | C14-GRR-NH2 | G | 40.53 | 31.94 (±1.99) | 8 (3.99) | 32 (1.00) | 16 (2.00) |
| 34 | C14-HRR-NH2 | H | 35.80 | 50.31 (±0.52) | 16 (3.14) | 64 (0.79) | 16 (3.14) |
| 37 | C12-IRR-NH2 | I | 39.05 | 120.40 (±1.55) | 8 (15.05) | 16 (7.53) | 16 (7.53) |
| 38 | C14-IRR-NH2 | I | 45.65 | 78.92 (±4.38) | 4 (19.73) | 4 (19.73) | 4 (19.73) |
| 42 | C14-KRR-NH2 | K | 35.66 | 212.08 (±6.12) | 8 (26.51) | 64 (3.31) | 32 (6.63) |
| 45 | C12-LRR-NH2 | L | 39.82 | 112.81 (±1.39) | 8 (14.10) | 16 (7.05) | 16 (7.05) |
| 46 | C14-LRR-NH2 | L | 46.44 | 29.50 (±1.05) | 4 (7.38) | 4 (7.38) | 4 (7.38) |
| 49 | C12-MRR-NH2 | M | 37.95 | 206.50 (±9.14) | 8 (25.81) | 32 (6.45) | 32 (6.45) |
| 50 | C14-MRR-NH2 | M | 44.69 | 35.11 (±0.90) | 4 (8.78) | 8 (4.39) | 4 (8.78) |
| 54 | C14-M(O)RR-NH2 | M(O) | 39.18 | 116.28 (±1.77) | 32 (3.63) | 64 (1.82) | 32 (3.63) |
| 58 | C14-M(O2)RR-NH2 | M(O2) | 41.17 | 70.11 (±2.93) | 16 (4.38) | 32 (2.19) | 16 (4.38) |
| 62 | C14-NRR-NH2 | N | 38.71 | 64.37 (±2.23) | 16 (4.02) | 32 (2.01) | 16 (4.02) |
| 64 | C10-NleRR-NH2 | Nle | 33.16 | >256 | 32 (>8) | 64 (>4) | 64 (>4) |
| 65 | C12-NleRR-NH2 | Nle | 39.96 | >256 | 8 (>32) | 8 (>32) | 16 (>16) |
| 66 | C14-NleRR-NH2 | Nle | 46.68 | 37.54 (±1.39) | 4 (9.39) | 4 (9.39) | 4 (9.39) |
| 69 | C12-NvaRR-NH2 | Nva | 37.67 | 207.62 (±5.27) | 8 (25.95) | 32 (6.49) | 32 (6.49) |
| 70 | C14-NvaRR-NH2 | Nva | 44.53 | 42.04 (±1.38) | 4 (10.51) | 8 (5.26) | 4 (10.51) |
| 73 | C12-PRR-NH2 | P | 35.82 | >256 | 32 (>8) | 64 (>4) | 64 (>4) |
| 74 | C14-PRR-NH2 | P | 42.61 | 53.41 (±1.84) | 16 (3.34) | 16 (3.34) | 8 (6.68) |
| 78 | C14-QRR-NH2 | Q | 38.52 | 72.31 (±3.00) | 16 (4.52) | 32 (2.26) | 32 (2.26) |
| 81 | C12-RRR-NH2 | R | 30.00 | >256 | 16 (>16) | 128 (>2) | 128 (>2) |
| 82 | C14-RRR-NH2 | R | 36.76 | 211.32 (±9.57) | 4 (52.83) | 32 (6.60) | 16 (13.21) |
| 86 | C14-SRR-NH2 | S | 39.72 | 41.54 (±1.29) | 16 (2.60) | 32 (1.30) | 32 (1.30) |
| 89 | C12-TRR-NH2 | T | 33.82 | >256 | 32 (>8) | 128 (>2) | 64 (>4) |
| 90 | C14-TRR-NH2 | T | 40.80 | 43.43 (±0.90) | 8 (5.43) | 32 (1.36) | 8 (5.43) |
| 93 | C12-VRR-NH2 | V | 37.17 | 246.30 (±14.12) | 16 (15.39) | 32 (7.70) | 32 (7.70) |
| 94 | C14-VRR-NH2 | V | 44.02 | 30.73 (±0.41) | 4 (7.68) | 8 (3.84) | 8 (3.84) |
| 96 | C10-WRR-NH2 | W | 34.31 | >256 | 16 (>16) | 32 (>8) | 32 (>8) |
| 97 | C12-WRR-NH2 | W | 40.79 | 42.42 (±0.57) | 4 (10.61) | 8 (5.30) | 8 (5.30) |
| 98 | C14-WRR-NH2 | W | 47.11 | 36.91 (±0.89) | 4 (9.23) | 8 (4.61) | 4 (9.23) |
| 101 | C12-YRR-NH2 | Y | 36.17 | 123.08 (±1.12) | 8 (15.39) | 16 (7.69) | 32 (3.85) |
| 102 | C14-YRR-NH2 | Y | 42.73 | 46.52 (±5.24) | 4 (11.63) | 8 (5.82) | 4 (11.63) |
| 105 | C10(6)-RR-NH2 | - | 39.62 | 200.25 (±16.07) | 4 (50.06) | 8 (25.03) | 16 (12.52) |
| Number of more selective analogs than reference lipopeptide | 22 | 34 | 20 | ||||
| Number of more selective and equal or more active analogs | 18 | 18 | 8 | ||||
| Code | Name | X | HC50 [µg/mL] | MIC [µg/mL] (SI) | ||||
|---|---|---|---|---|---|---|---|---|
| E1 | S | K | A | E2 | ||||
| 3 | C12-RR-NH2 | Reference compound | >256 | 32 (>8) | 32 (>8) | >256 (-) | 256 (>1.07) | >256 (-) |
| 4 | C14-RR-NH2 | 68.43 (±1.41) | 8 * (8.55) * | 8 * (8.55) * | 256 (0.27) | 64 (1.07) | 256 (0.27) | |
| 5 | C16-RR-NH2 | 22.91 (±1.18) | 8 (2.86) | 8 (2.86) | 32 (0.72) | 32 (0.72) | 32 (0.72) | |
| 6 | C18-RR-NH2 | 25.19 (±0.67) | 8 (3.15) | 16 (1.57) | 16 * (1.57) * | 32 * (0.79) * | 16 * (1.57) * | |
| 10 | C14-ARR-NH2 | A | 41.08 (±0.62) | 8 (5.14) | 16 (2.57) | 256 (0.16) | 32 (1.28) | 256 (0.16) |
| 14 | C14-C(Acm)RR-NH2 | C(Acm) | 39.72 (±1.25) | 8 (4.97) | 16 (2.48) | 256 (0.16) | 16 (2.48) | 256 (0.16) |
| 24 | C10-FRR-NH2 | F | >256 | 32 (>8) | 16 (>16) | >256 (-) | 64 (>4) | >256 (-) |
| 25 | C12-FRR-NH2 | F | 91.21 (±16.38) | 8 (11.40) | 8 (11.40) | 256 (0.36) | 64 (1.43) | 256 (0.36) |
| 26 | C14-FRR-NH2 | F | 39.09 (±1.18) | 16 (2.44) | 32 (1.22) | 64 (0.61) | 128 (0.31) | 64 (0.61) |
| 30 | C14-GRR-NH2 | G | 31.94 (±1.99) | 8 (3.99) | 8 (3.99) | 256 (0.12) | 64 (0.50) | 256 (0.12) |
| 34 | C14-HRR-NH2 | H | 50.31 (±0.52) | 8 (6.29) | 16 (3.14) | 256 (0.20) | 128 (0.39) | 256 (0.20) |
| 37 | C12-IRR-NH2 | I | 120.4 (±1.55) | 8 (15.05) | 8 (15.05) | 256 (0.47) | 64 (1.88) | 256 (0.47) |
| 38 | C14-IRR-NH2 | I | 78.92 (±4.38) | 4 (19.73) | 4 (19.73) | 64 (1.23) | 64 (1.23) | 64 (1.23) |
| 42 | C14-KRR-NH2 | K | 212.08 (±6.12) | 8 (26.51) | 8 (26.51) | 128 (1.66) | 64 (3.31) | 64 (3.31) |
| 45 | C12-LRR-NH2 | L | 112.81 (±1.39) | 16 (7.05) | 16 (7.05) | 256 (0.44) | 64 (1.76) | 256 (0.44) |
| 46 | C14-LRR-NH2 | L | 29.50 (±1.05) | 8 (3.69) | 16 (1.84) | 128 (0.23) | 64 (0.46) | 128 (0.23) |
| 49 | C12-MRR-NH2 | M | 206.5 (±9.14) | 16 (12.91) | 16 (12.91) | 256 (0.81) | 64 (3.23) | 256 (0.81) |
| 50 | C14-MRR-NH2 | M | 35.11 (±0.90) | 8 (4.39) | 8 (4.39) | 128 (0.27) | 32 (1.10) | 128 (0.27) |
| 54 | C14-M(O)RR-NH2 | M(O) | 116.28 (±1.77) | 16 (7.27) | 16 (7.27) | 256 (0.45) | 64 (1.82) | >256 (-) |
| 58 | C14-M(O2)RR-NH2 | M(O2) | 70.11 (±2.93) | 8 (8.76) | 16 (4.38) | 256 (0.27) | 32 (2.19) | 256 (0.27) |
| 62 | C14-NRR-NH2 | N | 64.37 (±2.23) | 8 (8.05) | 16 (4.02) | 128 (0.50) | 32 (2.01) | 128 (0.50) |
| 65 | C12-NleRR-NH2 | Nle | >256 | 8 (>32) | 8 (>32) | 256 (>1) | 32 (>8) | 256 (>1) |
| 66 | C14-NleRR-NH2 | Nle | 37.54 (±1.39) | 8 (4.69) | 8 (4.69) | 32 (1.17) | 64 (0.59) | 32 (1.17) |
| 69 | C12-NvaRR-NH2 | Nva | 207.62 (±5.27) | 16 (12.98) | 16 (12.98) | 256 (0.81) | 64 (3.24) | 256 (0.81) |
| 70 | C14-NvaRR-NH2 | Nva | 42.04 (±1.38) | 4 (10.51) | 4 (10.51) | 128 (0.33) | 64 (0.66) | 128 (0.33) |
| 74 | C14-PRR-NH2 | P | 53.41 (±1.84) | 8 (6.68) | 8 (6.68) | 256 (0.21) | 64 (0.83) | 256 (0.21) |
| 78 | C14-QRR-NH2 | Q | 72.31 (±3.00) | 16 (4.52) | 16 (4.52) | 256 (0.28) | 64 (1.13) | 256 (0.28) |
| 81 | C12-RRR-NH2 | R | >256 | 16 (>16) | 32 (>8) | >256 (-) | 256 (>1) | >256 (-) |
| 82 | C14-RRR-NH2 | R | 211.32 (±9.57) | 8 (26.42) | 8 (26.42) | 128 (1.65) | 128 (1.65) | 128 (1.65) |
| 86 | C14-SRR-NH2 | S | 41.54 (±1.29) | 16 (2.60) | 16 (2.60) | 128 (0.32) | 32 (1.30) | 128 (0.32) |
| 90 | C14-TRR-NH2 | T | 43.43 (±0.90) | 8 (5.43) | 8 (5.43) | 128 (0.34) | 64 (0.68) | 128 (0.34) |
| 93 | C12-VRR-NH2 | V | 246.3 (±14.12) | 16 (15.39) | 16 (15.39) | >256 (-) | 128 (1.92) | 256 (0.96) |
| 94 | C14-VRR-NH2 | V | 30.73 (±0.41) | 16 (1.92) | 4 (7.68) | 128 (0.24) | 32 (0.96) | 128 (0.24) |
| 96 | C10-WRR-NH2 | W | >256 | 16 (>16) | 16 (>16) | >256 (-) | 64 (>4) | >256 (-) |
| 97 | C12-WRR-NH2 | W | 42.42 (±0.57) | 8 (5.30) | 2 (21.21) | 256 (0.17) | 32 (1.33) | 256 (0.17) |
| 98 | C14-WRR-NH2 | W | 36.91 (±0.89) | 8 (4.61) | 2 (18.46) | 64 (0.58) | 128 (0.29) | 32 (1.15) |
| 101 | C12-YRR-NH2 | Y | 123.08 (±1.12) | 16 (7.69) | 8 (15.39) | 128 (0.96) | 32 (3.85) | 128 (0.96) |
| 102 | C12-YRR-NH2 | Y | 46.52 (±5.24) | 4 (11.63) | 4 (11.63) | 32 (1.45) | 32 (1.45) | 64 (0.73) |
| 105 | C10(6)-RR-NH2 | - | 200.25 (±16.07) | 8 (25.03) | 4 (50.06) | >256 (-) | 256 (0.78) | >256 (-) |
| Number of more selective compounds than reference lipopeptide | 15 | 17 | 2 | 26 | 2 | |||
| Number of more selective and equal or more active analogs | 10 | 12 | 0 | 11 | 0 | |||
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Neubauer, D.; Jaśkiewicz, M.; Bauer, M.; Gołacki, K.; Kamysz, W. Ultrashort Cationic Lipopeptides–Effect of N-Terminal Amino Acid and Fatty Acid Type on Antimicrobial Activity and Hemolysis. Molecules 2020, 25, 257. https://doi.org/10.3390/molecules25020257
Neubauer D, Jaśkiewicz M, Bauer M, Gołacki K, Kamysz W. Ultrashort Cationic Lipopeptides–Effect of N-Terminal Amino Acid and Fatty Acid Type on Antimicrobial Activity and Hemolysis. Molecules. 2020; 25(2):257. https://doi.org/10.3390/molecules25020257
Chicago/Turabian StyleNeubauer, Damian, Maciej Jaśkiewicz, Marta Bauer, Krzysztof Gołacki, and Wojciech Kamysz. 2020. "Ultrashort Cationic Lipopeptides–Effect of N-Terminal Amino Acid and Fatty Acid Type on Antimicrobial Activity and Hemolysis" Molecules 25, no. 2: 257. https://doi.org/10.3390/molecules25020257
APA StyleNeubauer, D., Jaśkiewicz, M., Bauer, M., Gołacki, K., & Kamysz, W. (2020). Ultrashort Cationic Lipopeptides–Effect of N-Terminal Amino Acid and Fatty Acid Type on Antimicrobial Activity and Hemolysis. Molecules, 25(2), 257. https://doi.org/10.3390/molecules25020257