The Structure and Function of Paraoxonase-1 and Its Comparison to Paraoxonase-2 and -3
Abstract
:1. Introduction
2. Paraoxonase 1: An Evolutionary Highlight with Many Enzymatic Activities
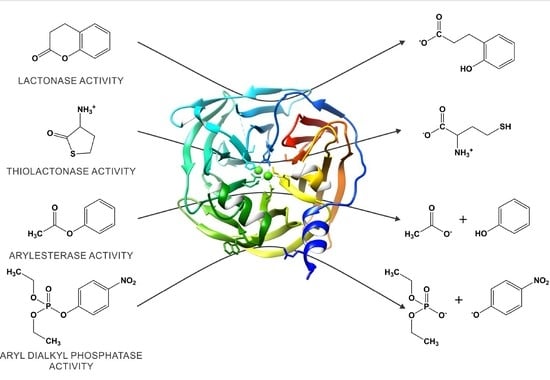
3. The Catalytic Versatility of PON1
4. Structural Insight into the Active Site of PON1
5. Paraoxonase 2: The Oldest Member of the Family
6. Paraoxonase 3: In the Shadow of his Younger Brother
7. Paraoxonase Versatility
8. Conclusions and Future Perspectives
- Structural data on PON2 and PON3 could explain the substrate preferences of all three PONs.
- Several studies have reported either higher or lower activity of predominantly PON1 in different diseases. However, it is still unclear whether elevated PON1 levels cause disease or represent a protective mechanism related to their antioxidative properties.
- The primary substrate of all three enzymes remains to be confirmed.
- Additional studies on point mutations could improve paraoxonase activity, which could enable the use of PON1 as an antidote in acute OP poisoning.
- Searching for PON agonists is of high importance in prevention strategies of development of cardiovascular diseases.
- Studies focused on testing already approved drugs and their effect on PON activity may explain the side effect and further improve the treatment of various conditions.
- Although PONs are relatively small proteins, their binding to membranes and HDLs could be studied using artificial membranes. As studies report that binding to membranes (also in the presence of detergents) can promote PON oligomerization, cryoelectron microscopy could help elucidate this phenomenon.
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| Ca1 | catalytic calcium |
| Ca2 | structural calcium |
| DHC | dihydrocoumarin |
| HCTL | homocysteine thiolactone |
| HDLs | high-density lipoproteins |
| hPON1 | human PON1 |
| hPON2 | human PON2 |
| hPON3 | human PON3 |
| kDa | kilodaltons |
| kcat | turnover number |
| KM | Michaelis constant |
| kOH | second-order rate constant for the hydroxide ion reaction with carbonyl groups of various substrates |
| LDLs | low-density lipoproteins |
| OP | organophosphate |
| PA | phenyl acetate |
| PDB | protein database |
| PON1 | paraoxonase 1 |
| PON1 | paraoxonase 1 gene |
| PON2 | paraoxonase 2 |
| PON2 | paraoxonase 2 gene |
| PON3 | paraoxonase 3 |
| PON3 | paraoxonase 3 gene |
| rePON1 | recombinant PON1 |
References
- Aharoni, A.; Gaidukov, L.; Yagur, S.; Toker, L.; Silman, I.; Tawfik, D.S. Directed evolution of mammalian paraoxonases PON1 and PON3 for bacterial expression and catalytic specialization. Proc. Natl. Acad. Sci. USA 2004, 101, 482–487. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mackness, M.I. ‘A’-esterases. Enzymes looking for a role? Biochem. Pharmacol. 1989, 38, 385–390. [Google Scholar] [CrossRef]
- Walker, C.H. The classification of esterases which hydrolyse organophosphates: Recent developments. Chem. Biol. Interact. 1993, 87, 17–24. [Google Scholar] [CrossRef]
- Draganov, D.I.; Stetson, P.L.; Watson, C.E.; Billecke, S.S.; La Du, B.N. Rabbit serum paraoxonase 3 (PON3) is a high density lipoprotein-associated lactonase and protects low density lipoprotein against oxidation. J. Biol. Chem. 2000, 275, 33435–33442. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lu, H.; Zhu, J.; Zang, Y.; Ze, Y.; Qin, J. Cloning, purification, and refolding of human paraoxonase-3 expressed in Escherichia coli and its characterization. Protein Expr. Purif. 2006, 46, 92–99. [Google Scholar] [CrossRef]
- Deakin, S.; Leviev, I.; Gomaraschi, M.; Calabresi, L.; Franceschini, G.; James, R.W. Enzymatically active paraoxonase-1 is located at the external membrane of producing cells and released by a high affinity, saturable, desorption mechanism. J. Biol. Chem. 2002, 277, 4301–4308. [Google Scholar] [CrossRef] [Green Version]
- Huang, Y.; Wu, Z.; Riwanto, M.; Gao, S.; Levison, B.S.; Gu, X.; Fu, X.; Wagner, M.A.; Besler, C.; Gerstenecker, G.; et al. Myeloperoxidase, paraoxonase-1, and HDL form a functional ternary complex. J. Clin. Investig. 2013, 123, 3815–3828. [Google Scholar] [CrossRef] [Green Version]
- Marsillach, J.; Mackness, B.; Mackness, M.; Riu, F.; Beltran, R.; Joven, J.; Camps, J. Immunohistochemical analysis of paraoxonases-1, 2, and 3 expression in normal mouse tissues. Free Radic. Biol. Med. 2008, 45, 146–157. [Google Scholar] [CrossRef]
- Navab, M.; Imes, S.S.; Hama, S.Y.; Hough, G.P.; Ross, L.A.; Bork, R.W.; Valente, A.J.; Berliner, J.A.; Drinkwater, D.C.; Laks, H.; et al. Monocyte transmigration induced by modification of low density lipoprotein in cocultures of human aortic wall cells is due to induction of monocyte chemotactic protein 1 synthesis and is abolished by high density lipoprotein. J. Clin. Investig. 1991, 88, 2039–2046. [Google Scholar] [CrossRef]
- Shih, D.M.; Gu, L.; Xia, Y.R.; Navab, M.; Li, W.F.; Hama, S.; Castellani, L.W.; Furlong, C.E.; Costa, L.G.; Fogelman, A.M.; et al. Mice lacking serum paraoxonase are susceptible to organophosphate toxicity and atherosclerosis. Nature 1998, 394, 284–287. [Google Scholar] [CrossRef]
- Marsillach, J.; Costa, L.G.; Furlong, C.E. Paraoxonase-1 and Early-Life Environmental Exposures. Ann. Glob. Health 2016, 82, 100–110. [Google Scholar] [CrossRef] [PubMed]
- Bacchetti, T.; Ferretti, G.; Sahebkar, A. The role of paraoxonase in cancer. Semin. Cancer Biol. 2019, 56, 72–86. [Google Scholar] [CrossRef] [PubMed]
- Seshadri, S.; Beiser, A.; Selhub, J.; Jacques, P.F.; Rosenberg, I.H.; D’Agostino, R.B.; Wilson, P.W.; Wolf, P.A. Plasma homocysteine as a risk factor for dementia and Alzheimer’s disease. N. Engl. J. Med. 2002, 346, 476–483. [Google Scholar] [CrossRef] [PubMed]
- Undas, A.; Perla, J.; Lacinski, M.; Trzeciak, W.; Kazmierski, R.; Jakubowski, H. Autoantibodies against N-homocysteinylated proteins in humans: Implications for atherosclerosis. Stroke 2004, 35, 1299–1304. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vos, E. Homocysteine levels, paraoxonase 1 (PON1) activity, and cardiovascular risk. JAMA 2008, 300, 168–169. [Google Scholar] [CrossRef]
- Billecke, S.; Draganov, D.; Counsell, R.; Stetson, P.; Watson, C.; Hsu, C.; La Du, B.N. Human serum paraoxonase (PON1) isozymes Q and R hydrolyze lactones and cyclic carbonate esters. Drug Metab. Dispos. Biol. Fate Chem. 2000, 28, 1335–1342. [Google Scholar]
- La Du, B.N. Structural and functional diversity of paraoxonases. Nat. Med. 1996, 2, 1186–1187. [Google Scholar] [CrossRef]
- Biggadike, K.; Angell, R.M.; Burgess, C.M.; Farrell, R.M.; Hancock, A.P.; Harker, A.J.; Irving, W.R.; Ioannou, C.; Procopiou, P.A.; Shaw, R.E.; et al. Selective plasma hydrolysis of glucocorticoid gamma-lactones and cyclic carbonates by the enzyme paraoxonase: An ideal plasma inactivation mechanism. J. Med. Chem. 2000, 43, 19–21. [Google Scholar] [CrossRef]
- Khersonsky, O.; Tawfik, D.S. Structure-reactivity studies of serum paraoxonase PON1 suggest that its native activity is lactonase. Biochemistry 2005, 44, 6371–6382. [Google Scholar] [CrossRef]
- Mackness, B.; Mackness, M.I.; Arrol, S.; Turkie, W.; Durrington, P.N. Effect of the molecular polymorphisms of human paraoxonase (PON1) on the rate of hydrolysis of paraoxon. Br. J. Pharmacol. 1997, 122, 265–268. [Google Scholar] [CrossRef] [Green Version]
- Garin, M.C.; James, R.W.; Dussoix, P.; Blanche, H.; Passa, P.; Froguel, P.; Ruiz, J. Paraoxonase polymorphism Met-Leu54 is associated with modified serum concentrations of the enzyme. A possible link between the paraoxonase gene and increased risk of cardiovascular disease in diabetes. J. Clin. Investig. 1997, 99, 62–66. [Google Scholar] [CrossRef] [Green Version]
- Perla-Kajan, J.; Jakubowski, H. Paraoxonase 1 and homocysteine metabolism. Amino Acids 2012, 43, 1405–1417. [Google Scholar] [CrossRef]
- Josse, D.; Lockridge, O.; Xie, W.; Bartels, C.F.; Schopfer, L.M.; Masson, P. The active site of human paraoxonase (Pon1). J. Appl. Toxicol. 2001, 21 (Suppl. S1), S7–S11. [Google Scholar] [CrossRef]
- Gupta, R.D.; Goldsmith, M.; Ashani, Y.; Simo, Y.; Mullokandov, G.; Bar, H.; Ben-David, M.; Leader, H.; Margalit, R.; Silman, I.; et al. Directed evolution of hydrolases for prevention of G-type nerve agent intoxication. Nat. Chem. Biol. 2011, 7, 120–125. [Google Scholar] [CrossRef] [PubMed]
- La Du, B.N.; Aviram, M.; Billecke, S.; Navab, M.; Primo-Parmo, S.; Sorenson, R.C.; Standiford, T.J. On the physiological role(s) of the paraoxonases. Chem. Biol. Interact. 1999, 119–120, 379–388. [Google Scholar] [CrossRef]
- Draganov, D.I.; Teiber, J.F.; Speelman, A.; Osawa, Y.; Sunahara, R.; La Du, B.N. Human paraoxonases (PON1, PON2, and PON3) are lactonases with overlapping and distinct substrate specificities. J. Lipid Res. 2005, 46, 1239–1247. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Teiber, J.F.; Draganov, D.I.; La Du, B.N. Lactonase and lactonizing activities of human serum paraoxonase (PON1) and rabbit serum PON3. Biochem. Pharmacol. 2003, 66, 887–896. [Google Scholar] [CrossRef]
- Harel, M.; Aharoni, A.; Gaidukov, L.; Brumshtein, B.; Khersonsky, O.; Meged, R.; Dvir, H.; Ravelli, R.B.; McCarthy, A.; Toker, L.; et al. Structure and evolution of the serum paraoxonase family of detoxifying and anti-atherosclerotic enzymes. Nat. Struct. Mol. Biol. 2004, 11, 412–419. [Google Scholar] [CrossRef]
- Khersonsky, O.; Tawfik, D.S. The histidine 115-histidine 134 dyad mediates the lactonase activity of mammalian serum paraoxonases. J. Biol. Chem. 2006, 281, 7649–7656. [Google Scholar] [CrossRef] [Green Version]
- Ben-David, M.; Elias, M.; Filippi, J.J.; Dunach, E.; Silman, I.; Sussman, J.L.; Tawfik, D.S. Catalytic versatility and backups in enzyme active sites: The case of serum paraoxonase 1. J. Mol. Biol. 2012, 418, 181–196. [Google Scholar] [CrossRef]
- Muthukrishnan, S.; Shete, V.S.; Sanan, T.T.; Vyas, S.; Oottikkal, S.; Porter, L.M.; Magliery, T.J.; Hadad, C.M. Mechanistic Insights into the Hydrolysis of Organophosphorus Compounds by Paraoxonase-1: Exploring the Limits of Substrate Tolerance in a Promiscuous Enzyme. J. Phys. Org. Chem. 2012, 25, 1247–1260. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lin, B.B.; Su, H.; Ma, G.C.; Liu, Y.J.; Hou, Q.Q. Theoretical study of the hydrolysis mechanism of dihydrocoumarin catalyzed by serum paraoxonase 1 (PON1): Different roles of Glu53 and His115 for catalysis. RSC Adv. 2016, 6, 60376–60384. [Google Scholar] [CrossRef]
- Debord, J.; Bollinger, J.C.; Harel, M.; Dantoine, T. Temperature dependence of binding and catalysis for human serum arylesterase/paraoxonase. Biochimie 2014, 97, 72–77. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bavec, A.; Knez, D.; Makovec, T.; Stojan, J.; Gobec, S.; Golicnik, M. Exploring the aryl esterase catalysis of paraoxonase-1 through solvent kinetic isotope effects and phosphonate-based isosteric analogues of the tetrahedral reaction intermediate. Biochimie 2014, 106, 184–186. [Google Scholar] [CrossRef] [PubMed]
- Izbicka, E.; Bolen, D.W. Hydrolysis Enthalpy Changes for Selected 5-Membered and 6-Membered Cyclic Esters. J. Am. Chem. Soc. 1978, 100, 7625–7628. [Google Scholar] [CrossRef]
- Dumas, D.P.; Caldwell, S.R.; Wild, J.R.; Raushel, F.M. Purification and properties of the phosphotriesterase from Pseudomonas diminuta. J. Biol. Chem. 1989, 264, 19659–19665. [Google Scholar]
- Purcell, J.; Hengge, A.C. The thermodynamics of phosphate versus phosphorothioate ester hydrolysis. J. Org. Chem. 2005, 70, 8437–8442. [Google Scholar] [CrossRef]
- Garel, J.; Tawfik, D.S. Mechanism of hydrolysis and aminolysis of homocysteine thiolactone. Chemistry 2006, 12, 4144–4152. [Google Scholar] [CrossRef]
- Kuo, C.L.; La Du, B.N. Calcium binding by human and rabbit serum paraoxonases. Structural stability and enzymatic activity. Drug Metab. Dispos. Biol. Fate Chem. 1998, 26, 653–660. [Google Scholar]
- Brushia, R.J.; Forte, T.M.; Oda, M.N.; La Du, B.N.; Bielicki, J.K. Baculovirus-mediated expression and purification of human serum paraoxonase 1A. J. Lipid Res. 2001, 42, 951–958. [Google Scholar]
- Josse, D.; Ebel, C.; Stroebel, D.; Fontaine, A.; Borges, F.; Echalier, A.; Baud, D.; Renault, F.; Le Maire, M.; Chabrieres, E.; et al. Oligomeric states of the detergent-solubilized human serum paraoxonase (PON1). J. Biol. Chem. 2002, 277, 33386–33397. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Josse, D.; Xie, W.; Renault, F.; Rochu, D.; Schopfer, L.M.; Masson, P.; Lockridge, O. Identification of residues essential for human paraoxonase (PON1) arylesterase/organophosphatase activities. Biochemistry 1999, 38, 2816–2825. [Google Scholar] [CrossRef] [PubMed]
- Aviram, M.; Billecke, S.; Sorenson, R.; Bisgaier, C.; Newton, R.; Rosenblat, M.; Erogul, J.; Hsu, C.; Dunlop, C.; La Du, B. Paraoxonase active site required for protection against LDL oxidation involves its free sulfhydryl group and is different from that required for its arylesterase/paraoxonase activities: Selective action of human paraoxonase allozymes Q and R. Arterioscler. Thromb. Vasc. Biol. 1998, 18, 1617–1624. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Otto, T.C.; Harsch, C.K.; Yeung, D.T.; Magliery, T.J.; Cerasoli, D.M.; Lenz, D.E. Dramatic differences in organophosphorus hydrolase activity between human and chimeric recombinant mammalian paraoxonase-1 enzymes. Biochemistry 2009, 48, 10416–10422. [Google Scholar] [CrossRef] [Green Version]
- Yeung, D.T.; Lenz, D.E.; Cerasoli, D.M. Analysis of active-site amino-acid residues of human serum paraoxonase using competitive substrates. FEBS J. 2005, 272, 2225–2230. [Google Scholar] [CrossRef]
- Ben-David, M.; Wieczorek, G.; Elias, M.; Silman, I.; Sussman, J.L.; Tawfik, D.S. Catalytic metal ion rearrangements underline promiscuity and evolvability of a metalloenzyme. J. Mol. Biol. 2013, 425, 1028–1038. [Google Scholar] [CrossRef]
- Ben-David, M.; Sussman, J.L.; Maxwell, C.I.; Szeler, K.; Kamerlin, S.C.L.; Tawfik, D.S. Catalytic stimulation by restrained active-site floppiness--the case of high density lipoprotein-bound serum paraoxonase-1. J. Mol. Biol. 2015, 427, 1359–1374. [Google Scholar] [CrossRef]
- Gu, X.; Huang, Y.; Levison, B.S.; Gerstenecker, G.; DiDonato, A.J.; Hazen, L.B.; Lee, J.; Gogonea, V.; DiDonato, J.A.; Hazen, S.L. Identification of Critical Paraoxonase 1 Residues Involved in High Density Lipoprotein Interaction. J. Biol. Chem. 2016, 291, 1890–1904. [Google Scholar] [CrossRef] [Green Version]
- Ben-David, M.; Soskine, M.; Dubovetskyi, A.; Cherukuri, K.P.; Dym, O.; Sussman, J.L.; Liao, Q.; Szeler, K.; Kamerlin, S.C.L.; Tawfik, D.S. Enzyme Evolution: An Epistatic Ratchet versus a Smooth Reversible Transition. Mol. Biol. Evol. 2020, 37, 1133–1147. [Google Scholar] [CrossRef]
- Giordano, G.; Tait, L.; Furlong, C.E.; Cole, T.B.; Kavanagh, T.J.; Costa, L.G. Gender differences in brain susceptibility to oxidative stress are mediated by levels of paraoxonase-2 expression. Free Radic. Biol. Med. 2013, 58, 98–108. [Google Scholar] [CrossRef] [Green Version]
- Levy, E.; Trudel, K.; Bendayan, M.; Seidman, E.; Delvin, E.; Elchebly, M.; Lavoie, J.C.; Precourt, L.P.; Amre, D.; Sinnett, D. Biological role, protein expression, subcellular localization, and oxidative stress response of paraoxonase 2 in the intestine of humans and rats. Am. J. Physiol. Gastrointest. Liver Physiol. 2007, 293, G1252–G1261. [Google Scholar] [CrossRef] [Green Version]
- Rosenblat, M.; Draganov, D.; Watson, C.E.; Bisgaier, C.L.; La Du, B.N.; Aviram, M. Mouse macrophage paraoxonase 2 activity is increased whereas cellular paraoxonase 3 activity is decreased under oxidative stress. Arterioscler. Thromb. Vasc. Biol. 2003, 23, 468–474. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mochizuki, H.; Scherer, S.W.; Xi, T.; Nickle, D.C.; Majer, M.; Huizenga, J.J.; Tsui, L.C.; Prochazka, M. Human PON2 gene at 7q21.3: Cloning, multiple mRNA forms, and missense polymorphisms in the coding sequence. Gene 1998, 213, 149–157. [Google Scholar] [CrossRef]
- Barathi, S.; Charanya, M.; Muthukumaran, S.; Angayarkanni, N.; Umashankar, V. Comparative modeling of PON2 and analysis of its substrate binding interactions using computational methods. J. Ocul. Biol. Dis. Inform. 2010, 3, 64–72. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Altenhofer, S.; Witte, I.; Teiber, J.F.; Wilgenbus, P.; Pautz, A.; Li, H.; Daiber, A.; Witan, H.; Clement, A.M.; Forstermann, U.; et al. One enzyme, two functions: PON2 prevents mitochondrial superoxide formation and apoptosis independent from its lactonase activity. J. Biol. Chem. 2010, 285, 24398–24403. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Devarajan, A.; Bourquard, N.; Hama, S.; Navab, M.; Grijalva, V.R.; Morvardi, S.; Clarke, C.F.; Vergnes, L.; Reue, K.; Teiber, J.F.; et al. Paraoxonase 2 deficiency alters mitochondrial function and exacerbates the development of atherosclerosis. Antioxid. Redox Signal. 2011, 14, 341–351. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hagmann, H.; Kuczkowski, A.; Ruehl, M.; Lamkemeyer, T.; Brodesser, S.; Horke, S.; Dryer, S.; Schermer, B.; Benzing, T.; Brinkkoetter, P.T. Breaking the chain at the membrane: Paraoxonase 2 counteracts lipid peroxidation at the plasma membrane. FASEB J. 2014, 28, 1769–1779. [Google Scholar] [CrossRef]
- Horke, S.; Witte, I.; Wilgenbus, P.; Kruger, M.; Strand, D.; Forstermann, U. Paraoxonase-2 reduces oxidative stress in vascular cells and decreases endoplasmic reticulum stress-induced caspase activation. Circulation 2007, 115, 2055–2064. [Google Scholar] [CrossRef]
- Ng, C.J.; Bourquard, N.; Grijalva, V.; Hama, S.; Shih, D.M.; Navab, M.; Fogelman, A.M.; Lusis, A.J.; Young, S.; Reddy, S.T. Paraoxonase-2 deficiency aggravates atherosclerosis in mice despite lower apolipoprotein-B-containing lipoproteins: Anti-atherogenic role for paraoxonase-2. J. Biol. Chem. 2006, 281, 29491–29500. [Google Scholar] [CrossRef] [Green Version]
- Li, W.; Kennedy, D.; Shao, Z.; Wang, X.; Kamdar, A.K.; Weber, M.; Mislick, K.; Kiefer, K.; Morales, R.; Agatisa-Boyle, B.; et al. Paraoxonase 2 prevents the development of heart failure. Free Radic. Biol. Med. 2018, 121, 117–126. [Google Scholar] [CrossRef]
- Teiber, J.F.; Horke, S.; Haines, D.C.; Chowdhary, P.K.; Xiao, J.; Kramer, G.L.; Haley, R.W.; Draganov, D.I. Dominant role of paraoxonases in inactivation of the Pseudomonas aeruginosa quorum-sensing signal N-(3-oxododecanoyl)-L-homoserine lactone. Infect. Immun. 2008, 76, 2512–2519. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ozer, E.A.; Pezzulo, A.; Shih, D.M.; Chun, C.; Furlong, C.; Lusis, A.J.; Greenberg, E.P.; Zabner, J. Human and murine paraoxonase 1 are host modulators of Pseudomonas aeruginosa quorum-sensing. FEMS Microbiol. Lett. 2005, 253, 29–37. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stoltz, D.A.; Ozer, E.A.; Recker, T.J.; Estin, M.; Yang, X.; Shih, D.M.; Lusis, A.J.; Zabner, J. A common mutation in paraoxonase-2 results in impaired lactonase activity. J. Biol. Chem. 2009, 284, 35564–35571. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mandrich, L.; Cerreta, M.; Manco, G. An Engineered Version of Human PON2 Opens the Way to Understand the Role of Its Post-Translational Modifications in Modulating Catalytic Activity. PLoS ONE 2015, 10, e0144579. [Google Scholar] [CrossRef]
- Mackness, B.; McElduff, P.; Mackness, M.I. The paraoxonase-2-310 polymorphism is associated with the presence of microvascular complications in diabetes mellitus. J. Intern. Med. 2005, 258, 363–368. [Google Scholar] [CrossRef]
- Marchegiani, F.; Spazzafumo, L.; Provinciali, M.; Cardelli, M.; Olivieri, F.; Franceschi, C.; Lattanzio, F.; Antonicelli, R. Paraoxonase2 C311S polymorphism and low levels of HDL contribute to a higher mortality risk after acute myocardial infarction in elderly patients. Mol. Genet. Metab. 2009, 98, 314–318. [Google Scholar] [CrossRef]
- Shi, J.; Zhang, S.; Tang, M.; Liu, X.; Li, T.; Han, H.; Wang, Y.; Guo, Y.; Zhao, J.; Li, H.; et al. Possible association between Cys311Ser polymorphism of paraoxonase 2 gene and late-onset Alzheimer’s disease in Chinese. Brain Res. Mol. Brain Res. 2004, 120, 201–204. [Google Scholar] [CrossRef]
- Shih, D.M.; Meng, Y.; Sallam, T.; Vergnes, L.; Shu, M.L.; Reue, K.; Tontonoz, P.; Fogelman, A.M.; Lusis, A.J.; Reddy, S.T. PON2 Deficiency Leads to Increased Susceptibility to Diet-Induced Obesity. Antioxidants 2019, 8, 19. [Google Scholar] [CrossRef] [Green Version]
- Shi, S.; Buck, T.M.; Kinlough, C.L.; Marciszyn, A.L.; Hughey, R.P.; Chalfie, M.; Brodsky, J.L.; Kleyman, T.R. Regulation of the epithelial Na(+) channel by paraoxonase-2. J. Biol. Chem. 2017, 292, 15927–15938. [Google Scholar] [CrossRef] [Green Version]
- Sulaiman, D.; Li, J.; Devarajan, A.; Cunningham, C.M.; Li, M.; Fishbein, G.A.; Fogelman, A.M.; Eghbali, M.; Reddy, S.T. Paraoxonase 2 protects against acute myocardial ischemia-reperfusion injury by modulating mitochondrial function and oxidative stress via the PI3K/Akt/GSK-3beta RISK pathway. J. Mol. Cell. Cardiol. 2019, 129, 154–164. [Google Scholar] [CrossRef]
- Nagarajan, A.; Dogra, S.K.; Sun, L.; Gandotra, N.; Ho, T.; Cai, G.; Cline, G.; Kumar, P.; Cowles, R.A.; Wajapeyee, N. Paraoxonase 2 Facilitates Pancreatic Cancer Growth and Metastasis by Stimulating GLUT1-Mediated Glucose Transport. Mol. Cell 2017, 67, 685–701.e6. [Google Scholar] [CrossRef] [PubMed]
- Devarajan, A.; Su, F.; Grijalva, V.; Yalamanchi, M.; Yalamanchi, A.; Gao, F.; Trost, H.; Nwokedi, J.; Farias-Eisner, G.; Farias-Eisner, R.; et al. Paraoxonase 2 overexpression inhibits tumor development in a mouse model of ovarian cancer. Cell Death Dis. 2018, 9, 392. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, J.H.; Lu, S.J.; Wu, P.; Kong, L.C.; Ning, C.; Li, H.Y. Molecular mechanism whereby paraoxonase-2 regulates coagulation activation through endothelial tissue factor in rat haemorrhagic shock model. Int. Wound J. 2020, 17, 735–741. [Google Scholar] [CrossRef]
- Schweikert, E.M.; Devarajan, A.; Witte, I.; Wilgenbus, P.; Amort, J.; Forstermann, U.; Shabazian, A.; Grijalva, V.; Shih, D.M.; Farias-Eisner, R.; et al. PON3 is upregulated in cancer tissues and protects against mitochondrial superoxide-mediated cell death. Cell Death Differ. 2012, 19, 1549–1560. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shih, D.M.; Xia, Y.R.; Yu, J.M.; Lusis, A.J. Temporal and tissue-specific patterns of Pon3 expression in mouse: In situ hybridization analysis. Adv. Exp. Med. Biol. 2010, 660, 73–87. [Google Scholar] [PubMed] [Green Version]
- Mizoi, K.; Takahashi, M.; Sakai, S.; Ogihara, T.; Haba, M.; Hosokawa, M. Structure-activity relationship of atorvastatin derivatives for metabolic activation by hydrolases. Xenobiotica Fate Foreign Compd. Biol. Syst. 2020, 50, 261–269. [Google Scholar] [CrossRef] [PubMed]
- Draganov, D.I. Lactonases with organophosphatase activity: Structural and evolutionary perspectives. Chem.-Biol. Interact. 2010, 187, 370–372. [Google Scholar] [CrossRef] [PubMed]
- Rodrigo, L.; Gil, F.; Hernandez, A.F.; Lopez, O.; Pla, A. Identification of paraoxonase 3 in rat liver microsomes: Purification and biochemical properties. Biochem. J. 2003, 376, 261–268. [Google Scholar] [CrossRef] [Green Version]
- Furlong, C.E.; Cole, T.B.; Pettan-Brewer, C.; Geiss, G.; Richter, R.; Shih, D.M.; Tward, A.; Lusis, A.J.; Costa, L.G. Genetic and temporal determinants of pesticide sensitivity in children: Role of paraoxonase (PON1). Neurotoxicology 2004, 25, 688–689. [Google Scholar]
- Garcia-Heredia, A.; Marsillach, J.; Aragones, G.; Guardiola, M.; Rull, A.; Beltran-Debon, R.; Folch, A.; Mackness, B.; Mackness, M.; Pedro-Botet, J.; et al. Serum paraoxonase-3 concentration is associated with the severity of hepatic impairment in patients with chronic liver disease. Clin. Biochem. 2011, 44, 1320–1324. [Google Scholar] [CrossRef] [PubMed]
- Rull, A.; Garcia, R.; Fernandez-Sender, L.; Garcia-Heredia, A.; Aragones, G.; Beltran-Debon, R.; Marsillach, J.; Alegret, J.M.; Martin-Paredero, V.; Mackness, B.; et al. Serum paraoxonase-3 concentration is associated with insulin sensitivity in peripheral artery disease and with inflammation in coronary artery disease. Atherosclerosis 2012, 220, 545–551. [Google Scholar] [CrossRef] [PubMed]
- Samouilidou, E.; Bountou, E.; Papandroulaki, F.; Papamanolis, M.; Papakostas, D.; Grapsa, E. Serum Endocan Levels are Associated with Paraoxonase 1 Concentration in Patients with Chronic Kidney Disease. Ther. Apher. Dial. 2018, 22, 325–331. [Google Scholar] [CrossRef] [PubMed]
- Shih, D.M.; Xia, Y.R.; Wang, X.P.; Wang, S.S.; Bourquard, N.; Fogelman, A.M.; Lusis, A.J.; Reddy, S.T. Decreased obesity and atherosclerosis in human paraoxonase 3 transgenic mice. Circ. Res. 2007, 100, 1200–1207. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shamir, R.; Hartman, C.; Karry, R.; Pavlotzky, E.; Eliakim, R.; Lachter, J.; Suissa, A.; Aviram, M. Paraoxonases (PONs) 1, 2, and 3 are expressed in human and mouse gastrointestinal tract and in Caco-2 cell line: Selective secretion of PON1 and PON2. Free Radic. Biol. Med. 2005, 39, 336–344. [Google Scholar] [CrossRef]
- Primo-Parmo, S.L.; Sorenson, R.C.; Teiber, J.; La Du, B.N. The human serum paraoxonase/arylesterase gene (PON1) is one member of a multigene family. Genomics 1996, 33, 498–507. [Google Scholar] [CrossRef]
- Sorenson, R.C.; Primo-Parmo, S.L.; Camper, S.A.; La Du, B.N. The genetic mapping and gene structure of mouse paraoxonase/arylesterase. Genomics 1995, 30, 431–438. [Google Scholar] [CrossRef]
- Furlong, C.E.; Marsillach, J.; Jarvik, G.P.; Costa, L.G. Paraoxonases-1, -2 and -3: What are their functions? Chem.-Biol. Interact. 2016, 259, 51–62. [Google Scholar] [CrossRef] [Green Version]
- Teiber, J.F.; Xiao, J.; Kramer, G.L.; Ogawa, S.; Ebner, C.; Wolleb, H.; Carreira, E.M.; Shih, D.M.; Haley, R.W. Identification of biologically active delta-lactone eicosanoids as paraoxonase substrates. Biochem. Biophys. Res. Commun. 2018, 505, 87–92. [Google Scholar] [CrossRef]
- Draganov, D.I.; La Du, B.N. Pharmacogenetics of paraoxonases: A brief review. Naunyn-Schmiedebergs Arch. Pharmacol. 2004, 369, 78–88. [Google Scholar] [CrossRef]
- Porntadavity, S.; Permpongpaiboon, T.; Sukketsiri, W. Human paraoxonase 2. EXCLI J. 2010, 9, 159–172. [Google Scholar]
- Chen, R.; Jiang, X.; Sun, D.; Han, G.; Wang, F.; Ye, M.; Wang, L.; Zou, H. Glycoproteomics analysis of human liver tissue by combination of multiple enzyme digestion and hydrazide chemistry. J. Proteome Res. 2009, 8, 651–661. [Google Scholar] [CrossRef] [PubMed]
- Liu, T.; Qian, W.J.; Gritsenko, M.A.; Camp, D.G., 2nd; Monroe, M.E.; Moore, R.J.; Smith, R.D. Human plasma N-glycoproteome analysis by immunoaffinity subtraction, hydrazide chemistry, and mass spectrometry. J. Proteome Res. 2005, 4, 2070–2080. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stevens, R.C.; Suzuki, S.M.; Cole, T.B.; Park, S.S.; Richter, R.J.; Furlong, C.E. Engineered recombinant human paraoxonase 1 (rHuPON1) purified from Escherichia coli protects against organophosphate poisoning. Proc. Natl. Acad. Sci. USA 2008, 105, 12780–12784. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ding, G.Y.; Zhu, X.D.; Ji, Y.; Shi, G.M.; Shen, Y.H.; Zhou, J.; Fan, J.; Sun, H.C.; Huang, C. Serum PON1 as a biomarker for the estimation of microvascular invasion in hepatocellular carcinoma. Ann. Transl. Med. 2020, 8, 204. [Google Scholar] [CrossRef]
- Yu, Z.; Ou, Q.; Chen, F.; Bi, J.; Li, W.; Ma, J.; Wang, R.; Huang, X. Evaluation of the prognostic value of paraoxonase 1 in the recurrence and metastasis of hepatocellular carcinoma and establishment of a liver-specific predictive model of survival. J. Transl. Med. 2018, 16, 327. [Google Scholar] [CrossRef]
- Kedage, V.; Muttigi, M.S.; Shetty, M.S.; Suvarna, R.; Rao, S.S.; Joshi, C.; Prakash, M. Serum paraoxonase 1 activity status in patients with liver disorders. Saudi J. Gastroenterol. 2010, 16, 79–83. [Google Scholar]
- Xu, G.Y.; Lv, G.C.; Chen, Y.; Hua, Y.C.; Zhu, S.M.; Yang, Y.D. Monitoring the level of serum paraoxonase 1 activity in liver transplantation patients. Hepatobiliary Pancreat. Dis. Int. HBPD INT 2005, 4, 178–181. [Google Scholar]
- Li, X.C.; Wang, C.; Mulchandani, A.; Ge, X. Engineering Soluble Human Paraoxonase 2 for Quorum Quenching. ACS Chem. Biol. 2016, 11, 3122–3131. [Google Scholar] [CrossRef]
- Cai, J.; Yuan, S.X.; Yang, F.; Tao, Q.F.; Yang, Y.; Xu, Q.G.; Wang, Z.G.; Yu, J.; Lin, K.Y.; Wang, Z.Y.; et al. Paraoxonase 3 inhibits cell proliferation and serves as a prognostic predictor in hepatocellular carcinoma. Oncotarget 2016, 7, 70045–70057. [Google Scholar] [CrossRef] [Green Version]
- Huang, D.; Wang, Y.; He, Y.; Wang, G.; Wang, W.; Han, X.; Sun, Y.; Lin, L.; Shan, B.; Shen, G.; et al. Paraoxonase 3 is involved in the multi-drug resistance of esophageal cancer. Cancer Cell Int. 2018, 18, 168. [Google Scholar] [CrossRef]
- Costa, L.G.; Giordano, G.; Furlong, C.E. Pharmacological and dietary modulators of paraoxonase 1 (PON1) activity and expression: The hunt goes on. Biochem. Pharmacol. 2011, 81, 337–344. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alim, Z.; Kilic, D.; Demir, Y. Some indazoles reduced the activity of human serum paraoxonase 1, an antioxidant enzyme: In vitro inhibition and molecular modeling studies. Arch. Physiol. Biochem. 2019, 125, 387–395. [Google Scholar] [CrossRef] [PubMed]
- Aviram, M.; Rosenblat, M.; Bisgaier, C.L.; Newton, R.S.; Primo-Parmo, S.L.; La Du, B.N. Paraoxonase inhibits high-density lipoprotein oxidation and preserves its functions. A possible peroxidative role for paraoxonase. J. Clin. Investig. 1998, 101, 1581–1590. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Demir, Y. The behaviour of some antihypertension drugs on human serum paraoxonase-1: An important protector enzyme against atherosclerosis. J. Pharm. Pharmacol. 2019, 71, 1576–1583. [Google Scholar] [CrossRef] [PubMed]
- Demir, Y.; Balci, N.; Gurbuz, M. Differential effects of selective serotonin reuptake inhibitors on paraoxonase-1 enzyme activity: An in vitro study. Comp. Biochem. Physiol. Part C Toxicol. Pharmacol. 2019, 226, 108608. [Google Scholar] [CrossRef] [PubMed]
- Demir, Y.; Koksal, Z. The inhibition effects of some sulfonamides on human serum paraoxonase-1 (hPON1). Pharmacol. Rep. 2019, 71, 545–549. [Google Scholar] [CrossRef] [PubMed]
- Isik, M.; Beydemir, S.; Demir, Y.; Durgun, M.; Turkes, C.; Nasir, A.; Necip, A.; Akkus, M. Benzenesulfonamide derivatives containing imine and amine groups: Inhibition on human paraoxonase and molecular docking studies. Int. J. Biol. Macromol. 2020, 146, 1111–1123. [Google Scholar] [CrossRef]






| Substrate | ΔH (kJ/mol) | kOH (M−1 s−1) | kcat (s−1) | KM (mM) | kcat/KM (M−1 s−1) |
|---|---|---|---|---|---|
| phenyl acetate | −97 a | 1.3 a | 700 b | 1.2 b | 580,000 |
| dihydrocoumarin | −105 a | 824 a | 150 b | 0.13 b | 1,150,000 |
| paraoxon | n.d. | 0.075 c | 4.8 b | 0.8 b | 6000 |
| PON1 | PON2 | PON3 | |
|---|---|---|---|
| UniProt id | P27169 (PON1_HUMAN) | Q15165 (PON2_HUMAN) | Q15166 (PON3_HUMAN) |
| gene locus | long arm of chromosome 7 (7q21.3-q22.1) [85] | ||
| 9 exons and 8 introns [85] | |||
| number of aminoacids | 355 insertion of K106 [86] | 354 | 354 |
| theoretical molecular weight | 39.7 kDa | 39.4 kDa | 39.6 kDa |
| size from SDS-PAGE | 43 kDa [89] | 40–43 kDa [90] | 40–45 kDa [4,5] |
| calcium-dependent glycoprotein | yes [89] | yes | yes [4,5] |
| hydrophobic N-terminal region | yes [28] | yes [57] | yes |
| confirmed N-glycosylation sites | N253, N324 [91] | N254, N323 [55,63,64] | N323 [92] |
| other post-translational modifications | n.d. | ubiquitination of K168 [64] | n.d. |
| 3D structure (PDB id) | 1V04 [28], 4HHO [46], 4HHQ [46], 4Q1U [47], 6G82 [49], 6GMU [49], 6H0A [49], 3SRG [30], 3SRE [30] | n.d. | n.d. |
| EC classification | 3.1.1.2 3.1.1.81 2.1.8.1 | 3.1.1.2 3.1.1.81 | 3.1.1.2 3.1.1.81 2.1.8.1 |
| protection against oxidative stress | yes [10] | yes [55,59] | yes [74,84] |
| arylesterase activity | yes [18] | yes very low [26,61] | yes low [26,76] |
| lactonase activity | yes [19] | yes low against DHC [26,61] | yes high [26,76] |
| organophosphate activity | yes [23,24] | no [26,61] | yes paraoxon only [26,76] |
| synthesis | produced in the liver [8] | ubiquitously expressed [50,51,52] | produced in the liver and to a lesser extent in the kidney [4] |
| location | mainly bound to HDLs in blood plasma in complex with myeloperoxidase [7] | intracellular enzyme on the membrane of mitochondria on the ER on the plasma membrane [55,56,57,58] | mainly bound to HDLs in blood plasma and mitochondria [4,74,75] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Taler-Verčič, A.; Goličnik, M.; Bavec, A. The Structure and Function of Paraoxonase-1 and Its Comparison to Paraoxonase-2 and -3. Molecules 2020, 25, 5980. https://doi.org/10.3390/molecules25245980
Taler-Verčič A, Goličnik M, Bavec A. The Structure and Function of Paraoxonase-1 and Its Comparison to Paraoxonase-2 and -3. Molecules. 2020; 25(24):5980. https://doi.org/10.3390/molecules25245980
Chicago/Turabian StyleTaler-Verčič, Ajda, Marko Goličnik, and Aljoša Bavec. 2020. "The Structure and Function of Paraoxonase-1 and Its Comparison to Paraoxonase-2 and -3" Molecules 25, no. 24: 5980. https://doi.org/10.3390/molecules25245980
APA StyleTaler-Verčič, A., Goličnik, M., & Bavec, A. (2020). The Structure and Function of Paraoxonase-1 and Its Comparison to Paraoxonase-2 and -3. Molecules, 25(24), 5980. https://doi.org/10.3390/molecules25245980