Proteomic Analysis of Saccharomyces cerevisiae Response to Oxidative Stress Mediated by Cocoa Polyphenols Extract
Abstract
:1. Introduction
2. Results
2.1. Comparative Proteomic Analysis
2.2. The GO Ontology Analysis
2.2.1. The GO Overrepresentation Test: Biological Process
2.2.2. Pathway
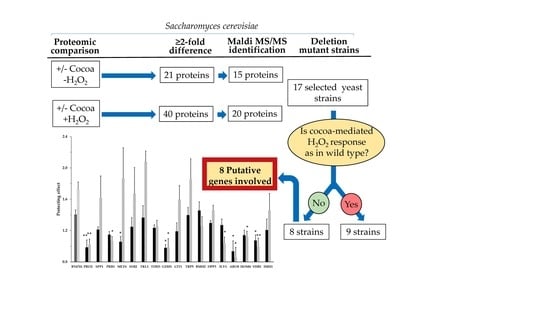
2.3. Antioxidant Response in Deletion Mutant Strains Potentially Involved in the Antioxidant Response of S. cerevisiae Mediated by Cocoa Extract
3. Discussion
4. Materials and Methods
4.1. Yeast Strains, Culture Media and Growth Conditions
4.2. Cocoa Polyphenol Extract
4.3. Sample Preparation for Proteomic Analysis
4.4. Protein Extraction and Two-Dimensional Gel Electrophoresis
4.5. Protein Visualization and Image Analysis
4.6. Protein Identification by MALDI-MS/MS
4.7. The GO Ontology Analysis
4.8. Assay to Evaluate the Antioxidant Response Induced by CPEX in the Deletion Mutant Strains
4.9. Statistical Analysis
Author Contributions
Funding
Conflicts of Interest
References
- Tomas-Barberán, F.A.; Cienfuegos-Jovellanos, E.; Marín, A.; Muguerza, B.; Gil-Izquierdo, A.; Cerdá, B.; Zafrilla, P.; Morillas, J.; Mulero, J.; Ibarra, A. A new process to develop a cocoa powder with higher flavonoid monomer content and enhanced bioavailability in healthy humans. J. Agric. Food Chem. 2007, 55, 3926–3935. [Google Scholar] [CrossRef]
- Lamuela-Raventos, R.M.; Romero-Pérez, A.I.; Andrés-La Cueva, C.; Tornero, A. Health effects of cocoa flavonoids. Food Sci. Tech. Int. 2005, 11, 159–176. [Google Scholar] [CrossRef]
- Martín, M.A.; Goya, L.; Ramos, S. Antidiabetic actions of cocoa flavanols. Mol. Nutr. Food Res. 2016, 60, 1756–1769. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Martín, M.A.; Ramos, S. Health beneficial effects of cocoa phenolic compounds: A mini-review. Curr. Opin. Food Sci. 2017, 14, 20–25. [Google Scholar] [CrossRef] [Green Version]
- Goya, L.; Martin, M.A.; Sarria, B.; Ramos, S.; Mateos, R.; Bravo, L. Effect of cocoa and its flavonoids on biomarkers of inflammation: Studies of cell culture, animals and humans. Nutrients 2016, 8, 212. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Teng, H.; Jia, Z.; Battino, M.; Miron, A.; Yu, Z.; Cao, H.; Xiao, J. Intracellular signaling pathways of inflammation modulated by dietary flavonoids: The most recent evidence. Crit. Rev. Food Sci. Nutr. 2018, 58, 2908–2924. [Google Scholar] [CrossRef] [PubMed]
- Chiva-Blanch, G.; Visioli, F. Polyphenols and health: Moving beyond antioxidants. J. Berry Res. 2012, 2, 63–71. [Google Scholar] [CrossRef] [Green Version]
- Edirisinghe, I.; Burton-Freeman, B. Anti-diabetic actions of berry polyphenols - review on proposed mechanisms of action. J. Berry Res. 2016, 6, 237–250. [Google Scholar] [CrossRef] [Green Version]
- Giampieri, F.; Alvarez-Suarez, J.M.; Battino, M. Strawberry and human health: Effects beyond antioxidant activity. J. Agric. Food Chem. 2014, 62, 3867–3876. [Google Scholar] [CrossRef]
- Giampieri, F.; Alvarez-Suarez, J.M.; Cordero, M.D.; Gasparrini, M.; Forbes-Hernández, T.Y.; Afrin, S.; Santos-Buelga, C.; González-Paramás, A.M.; Astolfi, P.; Rubini, C.; et al. Strawberry consumption improves aging-associated impairments, mitochondrial biogenesis and functionality through the AMP-activated protein kinase signaling cascade. Food Chem. 2017, 234, 464–471. [Google Scholar] [CrossRef]
- De La Torre-Ruiz, M.; Pujol, N.; Sundaran, V. Coping with oxidative stress. The yeast model. Curr. Drug Targets 2015, 16, 2–12. [Google Scholar] [CrossRef] [PubMed]
- Diezmann, S. Oxidative stress response and adaptation to H2O2 in the model eukaryote Saccharomyces cerevisiae and its human pathogenic relatives Candida albicans and Candida glabrata. Fungal Biol. Rev. 2014, 28, 126–136. [Google Scholar] [CrossRef]
- Goffeau, A.; Barrell, B.G.; Bussey, H.; Davis, R.W.; Dujon, B.; Feldmann, H.; Galibert, F.; Hoheisel, J.D.; Jacq, C.; Johnston, M.; et al. Life with 6000 genes. Science 1996, 274, 546–567. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Meng, D.; Zhang, P.; Li, S.; Ho, C.; Zhao, H. Antioxidant activity evaluation of dietary phytochemicals using Saccharomyces cerevisiae as a model. J. Funct. Foods 2017, 38, 36–44. [Google Scholar] [CrossRef]
- Mi, H.; Huang, X.; Muruganujan, A.; Tang, H.; Mills, C.; Kang, D.; Thomas, P.D. PANTHER version 11: Expanded annotation data fromGene Ontology and Reactome pathways, and dataanalysis tool enhancements. Nucleic Acids Res. 2017. [Google Scholar] [CrossRef] [Green Version]
- Martorell, P.; Forment, J.V.; de Llanos, R.; Montόn, F.; Llopis, S.; Gonzélez, N.; Genovés, S.; Cienfuegos, E.; Monzό, H.; Ramόn, D. Use of Saccharomyces cerevisiae and Caenorhabditis elegans as model organisms to study the effect of cocoa polyphenols in the resistance to oxidative stress. J. Agric. Food Chem. 2011, 59, 2071–2085. [Google Scholar] [CrossRef]
- Peláez-Soto, A.; Fernández-Espinar, M.T.; Roig, P.; Gil, J.V. Evaluation of the ability of polyphenol extracts of cocoa and red grape to promote the antioxidant response in yeast using a rapid multiwell assay. J. Food Sci. 2017, 82, 324–332. [Google Scholar] [CrossRef]
- Rajendran, P.; Nandakumar, N.; Rengarajan, T.; Palaniswami, R.; Gnanadhas, E.N.; Lakshminarasaiah, U.; Gopas, J.; Nishigaki, I. Antioxidants and human diseases. Clin. Chim. Acta 2014, 436, 332–347. [Google Scholar] [CrossRef]
- Katayama, S.; Mine, Y. Antioxidative activity of amino acids on tissue oxidative stress in human intestinal epithelial cell model. J. Agric. Food Chem. 2007, 55, 8458–8464. [Google Scholar] [CrossRef]
- Meister, A.; Anderson, M.E. Glutathione. Ann. Rev. Biochem. 1983, 52, 711–760. [Google Scholar] [CrossRef]
- Valerio, A.; D’Antona, G.; Nisoli, E. Branched-chain amino acids, mitochondrial biogenesis, and healthspan: An evolutionary perspective. Aging 2011, 3, 464–478. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shima, J.; Takagi, H. Stress-tolerance of baker’s-yeast (Saccharomyces cerevisiae) cells: Stress-protective molecules and genes involved in stress tolerance. Biotechnol. Appl. Biochem. 2009, 53, 155–164. [Google Scholar] [CrossRef] [PubMed]
- Liang, X.; Dickman, M.B.; Becker, D.F. Proline biosynthesis is required for endoplasmic reticulum stress tolerance in Saccharomyces cerevisiae. J. Biol. Chem. 2014, 289, 27794–27806. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guillamón, J.M.; van Riel, N.A.; Giuseppin, M.L.; Verrips, C. The glutamate synthase (GOGAT) of Saccharomyces cerevisiae plays an important role in central nitrogen metabolism. FEMS Yeast Res. 2001, 1, 169–175. [Google Scholar] [CrossRef]
- Jin, J.; Li, D.; Alesi, G.N.; Fan, J.N.; Kang, H.B.; Lu, Z.; Boggon, T.J.; Jin, P.; Yi, H.; Wright, E.R.; et al. Glutamate dehydrogenase 1 signals through antioxidant glutathione peroxidase 1 to regulate redox homeostasis and tumor growth. Cancer Cell 2015, 9, 257–270. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yelamanchi, S.D.; Jayaram, S.; Thomas, J.K.; Gundimeda, S.; Khan, A.A.; Singhal, A.; Keshava, P.T.S.; Pandey, A.; Somani, B.L.; Gowda, H. A pathway map of glutamate metabolism. J. Cell Commun. Signal. 2015, 3, 1–7. [Google Scholar] [CrossRef] [Green Version]
- Spanaki, C.; Zaganas, I.; Kounoupa, Z.; Plaitakis, A. The complex regulation of human glud1 and glud2 glutamate dehydrogenases and its implications in nerve tissue biology. Neurochem. Int. 2012, 61, 470–481. [Google Scholar] [CrossRef]
- Plaitakis, A.; Kalef-Ezra, E.; Kotzamani, D.; Zaganas, I.; Spanaki, C. The glutamate dehydrogenase pathway and its roles in cell and tissue biology in health and disease. Biology 2017, 6, 11. [Google Scholar] [CrossRef] [Green Version]
- Ohashi, K.; Chaleckis, R.; Takaine, M.; Wheelock, C.E.; Yoshida, S. Kynurenine aminotransferase activity of Aro8/Aro9 engage tryptophan degradation by producing kynurenic acid in Saccharomyces cerevisiae. Sci. Rep. 2017, 7, 12180. [Google Scholar] [CrossRef] [Green Version]
- Pérez-González, A.; Muñoz-Rugeles, L.; Alvarez-Idaboy, J.R. Tryptophan: Antioxidant or target of oxidative stress? A quantum chemistry elucidation. RSC Adv. 2014, 4, 56128–56131. [Google Scholar] [CrossRef]
- Petti, A.A.; Crutchfield, C.A.; Rabinowitz, J.D.; Botstein, D. Survival of starving yeast is correlated with oxidative stress response and nonrespiratory mitochondrial function. Proc. Natl. Acad. Sci. USA 2011, 108, 1089–E1098. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lafaye, A.; Junot, C.; Pereira, Y.; Lagniel, G.; Tabet, J.C.; Ezan, E.; Labarre, J. Combined proteome and metabolite-profiling analyses reveal surprising insights into yeast sulfur metabolism. J. Biol. Chem. 2005, 280, 24723–24730. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, L.; Zeng, M.; Hausladen, A.; Heitman, J.; Stamler, J.S. Protection from nitrosative stress by yeast flavohemoglobin. Proc. Natl. Acad. Sci. USA 2000, 97, 4672–4676. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lewinska, A.; Grzelak, A.; Bartosz, G. Application of a YHB1-GFP reporter to detect nitrosative stress in yeast. Red. Rep. 2008, 13, 161–171. [Google Scholar] [CrossRef]
- Kleinknecht, A.; Popova, B.; Lázaro, D.F.; Pinho, R.; Valerius, O.; Outeiro, T.F.; Braus, G.H. C-Terminal tyrosine residue modifications modulate the protective phosphorylation of serine 129 of α-synuclein in a yeast model of Parkinson’s disease. PLoS Genet. 2016, 12, e1006098. [Google Scholar] [CrossRef]
- Xue, Y.; Vashisht, A.A.; Tan, Y.; Su, T.; Wohlschlegel, J.A. PRB1 Is required for clipping of the histone h3 n terminal tail in Saccharomyces cerevisiae. PLoS ONE 2014, 9, e90496. [Google Scholar] [CrossRef] [Green Version]
- Meas, R.; Smerdon, M.J.; Wyrick, J.J. The amino-terminal tails of histones H2A and H3 coordinate efficient base excision repair, DNA damage signaling and postreplication repair in Saccharomyces cerevisiae. Nucleic Acids Res. 2015, 43, 4990–5001. [Google Scholar] [CrossRef] [Green Version]
- Yi, S.J.; Kimm, K. Histone tail cleavage as a novel epigenetic regulatory mechanism for gene expression. BMB Rep. 2018, 51, 211–218. [Google Scholar] [CrossRef] [Green Version]
- Joos, J.P.; Saadatmand, A.R.; Schnabel, C.; Viktorinová, I.; Brand, T.; Kramer, M.; Nattel, S.; Dobrev, D.; Tomancak, P.; Backs, J.; et al. Ectopic expression of S28A-mutated Histone H3 modulates longevity, stress resistance and cardiac function in Drosophila. Sci. Rep. 2018, 8, 2940. [Google Scholar] [CrossRef] [Green Version]
- Delaney, K.; Maillerm, J.; Wendam, J.M.; Gabusm, C.; Steiner, F.A. Differential expression of histone h3.3 genes and their role in modulating temperature stress response in Caenorhabditis elegans. Genetics 2018, 209, 551–565. [Google Scholar] [CrossRef] [Green Version]
- Su, X.B.; Pillus, L. Functions for diverse metabolic activities in heterochromatin. Proc. Natl. Acad. Sci. USA. 2016, 113, E1526–E1535. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ralser, M.; Wamelink, M.M.; Kowald, A.; Gerisch, B.; Heeren, G.; Struys, E.A.; Klipp, E.; Jakobs, C.; Breitenbach, M.; Lehrach, H.; et al. Dynamic rerouting of the carbohydrate flux is key to counteracting oxidative stress. J. Biol. 2007, 6, 10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Singleton, V.L.; Rossi, J.A. Colorimetry of total phenolics with phos-phomolybdic-phosphotungstic acid reagents. Am. J. Enol. Vitic. 1965, 16, 144–158. [Google Scholar]
- Gómez-Pastor, R.; Pérez-Torrado, R.; Cabiscol, E.; Matallana, E. Transcriptomic and proteomic insights of the wine yeast biomass propagation process. FEMS Yeast Res. 2010, 10, 870–884. [Google Scholar] [CrossRef] [Green Version]
- Chiari, M.; Nesi, M.; Roncada, P.; Righetti, P.G. Preparative Isoelectric Focusing in Multicompartment Electrolyzers: Novel, Hydrolytically Stable and Hydrophilic Isoelectric Membranes. Electrophoresis 1994, 15, 953–959. [Google Scholar] [CrossRef]
- Shevchenko, A.; Tomas, H.; Havlis, J.; Olsen, J.V.; Mann, M. In-gel digestion for mass spectrometric characterization of proteins and proteomes. Nat. Protoc. 2006, 1, 2856. [Google Scholar] [CrossRef]
- Suckau, D.; Resemann, A.; Schuerenberg, M.; Hufnagel, P.; Franzen, J.; Holle, A. A novel MALDI LIFT-TOF/TOF mass spectrometer for proteomics. Anal. Bioanal. Chem. 2003, 376, 952. [Google Scholar] [CrossRef]
- Perkins, D.N.; Pappin, D.J.; Creasy, D.M.; Cottrell, J.S. Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis 1999, 18, 3551–3567. [Google Scholar] [CrossRef]
Sample Availability: Samples of the compounds are available from the authors. |


| Spot | Protein | Protein Name | UnitProt Accession No. | Mass (Da) | Mascot Score 1 | Matched Peptides 2 | %COV 3 | Fold 4 |
|---|---|---|---|---|---|---|---|---|
| Without Stress Conditions | ||||||||
| 2601 | Cit1 | Citrate synthase, mitochondrial | P00890 | 53384 | 78 | 15 | 31 | +2.56 |
| 1703 | Ilv1 | Threonine dehydratase, mitochondrial | P00927 | 64076 | 96 | 18 | 38 | +2.40 |
| 3808 | Trp5 | Tryptophan synthase | P00931 | 76977 | 82 | 24 | 38 | +2.26 |
| 3902 | Eft1 5 | Elongation factor 2 | P32324 | 93686 | 255 | 37 | 42 | +2.38 |
| 6808 | Ssb2 | Ribosome-associated molecular chaperone SSB2 | P40150 | 66668 | 163 | 20 | 33 | +2.09 |
| 3908 | Met6 | 5-methyltetrahydropteroyltriglutamate--homocysteine methyltransferase | P05694 | 85978 | 115 | 31 | 41 | −2.03 |
| 5401 | Eft1 5 | Elongation factor 2 | P32324 | 93686 | 73 | 20 | 20 | −2.08 |
| 2707 | Cdc19 | Pyruvate kinase 1 | P00549 | 54909 | 258 | 32 | 59 | −2.22 |
| 4405 | Yhb1 | Flavohemoprotein | P39676 | 44846 | 165 | 17 | 57 | −2.34 |
| 3303 | Tdh3 | Glyceraldehyde-3-phosphate dehydrogenase 3 | P00359 | 35838 | 170 | 14 | 48 | −2.42 |
| 3301 | Rpl5 | 60S ribosomal protein L5 | P26321 | 33751 | 237 | 12 | 48 | −2.84 |
| 3502 | Pgk1 | Phosphoglycerate kinase | P00560 | 44768 | 273 | 23 | 53 | −2.85 |
| 6702 | Frs2 | Phenylalanine--tRNA ligase alpha subunit | P15625 | 57532 | 62 | 8 | 16 | −3.86 |
| 6501 | Eno2 | Enolase 2 | P00925 | 46942 | 131 | 11 | 29 | −3.89 |
| 3412 | Ilv5 | Ketol-acid reductoisomerase, mitochondrial | P06168 | 44512 | 129 | 17 | 43 | −4.90 |
| Under Oxidative Stress Conditions | ||||||||
| 7704 | Tkl1 | Transketolase 1 | P23254 | 73874 | 400 | 27 | 35 | +9.83 |
| 0905 | Cdc48 | Cell division control protein 48 | P25694 | 92167 | 382 | 38 | 44 | +5.38 |
| 6302 | Pgk1 6 | Phosphoglycerate kinase | P00560 | 44768 | 89 | 13 | 37 | +3.71 |
| 2612 | Pro2 | Gamma-glutamyl phosphate reductase | P54885 | 49881 | 125 | 16 | 46 | +2.91 |
| 4805 | YOL057W | Probable dipeptidyl peptidase 3 | Q08225 | 80745 | 89 | 24 | 35 | +2.87 |
| 4606 | Spp1 | COMPASS component SPP1 | Q03012 | 42468 | 61 | 15 | 38 | +2.48 |
| 1311 | Aim41 | Altered inheritance of mitochondria protein 41, mitochondrial | Q12032 | 21215 | 56 | 1 | 39 | −2.11 |
| 1222 | Sec14 | SEC14 cytosolic factor | P24280 | 35107 | 52 | 6 | 19 | −2.29 |
| 1108 | Tif1 | ATP-dependent RNA helicase eIF4A | P10081 | 44840 | 121 | 12 | 26 | −2.44 |
| 6801 | Met6 | 5-methyltetrahydropteroyltriglutamate--homocysteine methyltransferase | P05694 | 85978 | 79 | 23 | 34 | −2.98 |
| 1109 | Imh1 | Golgin IMH1 | Q06704 | 105333 | 64 | 34 | 36 | −3.28 |
| 3107 | Hom6 | Homoserine dehydrogenase | P31116 | 38478 | 421 | 19 | 35 | −4.59 |
| 2002 | Bmh2 | Protein BMH2 | P34730 | 31099 | 136 | 11 | 36 | −4.76 |
| 5411 | Aro8 | Aromatic/aminoadipate aminotransferase 1 | P53090 | 56371 | 181 | 9 | 14 | −4.77 |
| 6403 | Pgk1 6 | Phosphoglycerate kinase | P00560 | 44768 | 231 | 20 | 47 | −5.98 |
| 3605 | Gdh1 | NADP-specific glutamate dehydrogenase 1 | P07262 | 49881 | 142 | 17 | 38 | −7.47 |
| 8508 | Tef4 | Elongation factor 1-gamma 2 | P36008 | 46605 | 99 | 10 | 26 | −8.43 |
| 3510 | Eft1 | Elongation factor 2 | P32324 | 93686 | 130 | 24 | 25 | −10.91 |
| 8308 | Cdc19 | Pyruvate kinase 1 | P00549 | 54909 | 352 | 23 | 40 | −15.63 |
| 6205 | Prb1 | Cerevisin | P09232 | 69807 | 57 | 8 | 9 | −16.38 |
| GO Category Enriched (Biological Process) | Proteins Involved 1 | Observed Proteins | Expected Proteins | Fold Enrichment | P-Value 2 | Q-Value 3 |
|---|---|---|---|---|---|---|
| Cocoa Extract Effect without Oxidative Stress | ||||||
| isoleucine biosynthetic process | Ilv1, Ilv5 | 2 | 0.02 | 96.03 | 2.62 × 10−4 | 1.68 × 10−2 |
| alpha-amino acid biosynthetic process | Ilv1, Ilv5, Cit1, Met6, Trp5 | 5 | 0.26 | 19.52 | 4.00 × 10−6 | 7.10 × 10−4 |
| cellular amino acid metabolic process | Ilv1, Ilv5, frs2, Cit1, Met6, Trp5 | 6 | 0.55 | 10.91 | 8.98 × 10−6 | 1.29 × 10−3 |
| cellular amino acid biosynthetic process | Ilv1, Ilv5, Cit1, Met6, Trp5 | 5 | 0.27 | 18.33 | 5.39 × 10−6 | 8.45 × 10−4 |
| carboxylic acid biosynthetic process | Ilv1, Ilv5, Eno2, Cit1, Met6, Pgk1, Trp5, Tdh3, Cdc19 | 9 | 0.45 | 19.91 | 7.96 × 10−11 | 1.41 × 10−7 |
| branched-chain amino acid biosynthetic process | Ilv1, Ilv5 | 2 | 0.04 | 56.49 | 6.72 × 10−4 | 3.77 × 10−2 |
| glycolytic process | Eno2, Pgk1, Tdh3, Cdc19 | 4 | 0.05 | 73.87 | 3.10 × 10−7 | 1.50 × 10−4 |
| nicotinamide nucleotide biosynthetic process | Eno2, Pgk1, Tdh3, Cdc19 | 4 | 0.09 | 44.66 | 1.98 × 10−6 | 4.22 × 10−4 |
| pyruvate biosynthetic process | Eno2, Pgk1, Tdh3, Cdc19 | 4 | 0.05 | 73.87 | 3.10 × 10−7 | 1.84 × 10−4 |
| ATP biosynthetic process | Eno2, Pgk1, Tdh3, Cdc19 | 4 | 0.10 | 40.01 | 2.99 × 10−6 | 5.68 × 10−4 |
| nucleotide catabolic process | Eno2, Pgk1, Tdh3, Cdc19 | 4 | 0.07 | 60.02 | 6.62 × 10−7 | 2.35 × 10−4 |
| gluconeogenesis | Eno2, Pgk1, Tdh3 | 3 | 0.04 | 72.02 | 1.24 × 10−5 | 1.69 × 10−3 |
| reactive oxygen species metabolic process | Yhb1, Tdh3 | 2 | 0.04 | 50.54 | 8.24 × 10−4 | 4.39 × 10−2 |
| Cocoa Extract Effect under Oxidative Stress | ||||||
| isoleucine biosynthetic process | Hom6 | 1 | 0.03 | 37.34 | 2.90 × 10−2 | 1.18 |
| alpha-amino acid biosynthetic process | Hom6, Met6, Gdh1, Pro2, Aro8 | 5 | 0.33 | 15.18 | 1.61 × 10−5 | 1.71 × 10−2 |
| cellular amino acid metabolic process | Hom6, Met6, Gdh1, Pro2, Aro8 | 5 | 0.71 | 7.07 | 5.45 × 10−4 | 1.53 × 10−1 |
| cellular amino acid biosynthetic process | Hom6, Met6, Gdh1, Pro2, Aro8 | 5 | 0.35 | 14.25 | 2.16 × 10−5 | 1.91 × 10−2 |
| carboxylic acid metabolic process | Hom6, Met6, Gdh1, Pgk1 Pro2, Aro8, Cdc19 | 7 | 1.16 | 6.04 | 8.15 × 10−5 | 5.43 × 10−2 |
| branched-chain amino acid metabolic process | Hom6 | 1 | 0.08 | 12.45 | 7.97 × 10−2 | 1.84 |
| glycolytic process | Pgk1, Cdc19 | 2 | 0.07 | 28.73 | 2.44 × 10−3 | 3.52 × 10−1 |
| nicotinamide nucleotide biosynthetic process | Pgk1, Cdc19, tkl1 | 3 | 0.22 | 13.61 | 1.41 × 10−3 | 2.59 × 10−1 |
| pyruvate biosynthetic process | Pgk1, Cdc19 | 2 | 0.07 | 28.73 | 2.44 × 10−3 | 3.72 × 10−1 |
| ATP biosynthetic process | Pgk1, Cdc19 | 2 | 0.13 | 15.56 | 7.65 × 10−3 | 6.79 × 10−1 |
| nucleotide biosynthetic process | Pgk1, Cdc19 | 2 | 0.37 | 5.45 | 5.21 × 10−2 | 1.52 |
| gluconeogenesis | Pgk1 | 1 | 0.05 | 18.67 | 5.47 × 10−2 | 1.53 |
| Pathway | No. of Proteins | Proteins 1 | % 2 | % 3 |
|---|---|---|---|---|
| Without Oxidative Stress | ||||
| Apoptosis signaling pathway (P00006) | 1 | Ssb2 | 7.1 | 7.7 |
| Glycolysis (P00024) | 3 | Eno2, Pgk1, Tdh3 | 21.4 | 23.1 |
| Huntington disease (P00029) | 1 | Tdh3 | 7.1 | 7.7 |
| Isoleucine biosynthesis (P02748) | 2 | Ilv1. Ilv5 | 14.3 | 15.4 |
| Parkinson disease(P00049) | 1 | Ssb2 | 7.1 | 7.7 |
| Pyruvate metabolism (P02772) | 2 | Cit1. Cdc19 | 14.3 | 15.4 |
| TCA cycle (P00051) | 1 | Cit1 | 7.1 | 7.7 |
| Tryptophan biosynthesis (P02783): | 1 | Trp5 | 7.1 | 7.7 |
| Valine biosynthesis (P02785) | 1 | Ilv5 | 7.1 | 7.7 |
| With Oxidative Stress | ||||
| EGF receptor signaling pathway (P00018) | 1 | Bmh2 | 5.3 | 10 |
| GF signaling pathway (P00021) | 1 | Bmh2 | 5.3 | 10 |
| Glutamine glutamate conversion (P02745) | 1 | Gdh1 | 5.3 | 10 |
| Glycolysis (P00024) | 1 | Pgk1 | 5.3 | 10 |
| Lysine biosynthesis (P02751) | 1 | Hom6 | 5.3 | 10 |
| Parkinson disease (P00049) | 1 | Bmh2 | 5.3 | 10 |
| Pentose phosphate pathway (P02762) | 1 | Tkl1 | 5.3 | 10 |
| Proline biosynthesis (P02768) | 1 | Pro2 | 5.3 | 10 |
| Pyruvate metabolism (P02772) | 1 | Cdc19 | 5.3 | 10 |
| Threonine biosynthesis (P02781) | 1 | Hom6 | 5.3 | 10 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Peláez-Soto, A.; Roig, P.; Martínez-Culebras, P.V.; Fernández-Espinar, M.T.; Gil, J.V. Proteomic Analysis of Saccharomyces cerevisiae Response to Oxidative Stress Mediated by Cocoa Polyphenols Extract. Molecules 2020, 25, 452. https://doi.org/10.3390/molecules25030452
Peláez-Soto A, Roig P, Martínez-Culebras PV, Fernández-Espinar MT, Gil JV. Proteomic Analysis of Saccharomyces cerevisiae Response to Oxidative Stress Mediated by Cocoa Polyphenols Extract. Molecules. 2020; 25(3):452. https://doi.org/10.3390/molecules25030452
Chicago/Turabian StylePeláez-Soto, Ana, Patricia Roig, Pedro Vicente Martínez-Culebras, María Teresa Fernández-Espinar, and José Vicente Gil. 2020. "Proteomic Analysis of Saccharomyces cerevisiae Response to Oxidative Stress Mediated by Cocoa Polyphenols Extract" Molecules 25, no. 3: 452. https://doi.org/10.3390/molecules25030452
APA StylePeláez-Soto, A., Roig, P., Martínez-Culebras, P. V., Fernández-Espinar, M. T., & Gil, J. V. (2020). Proteomic Analysis of Saccharomyces cerevisiae Response to Oxidative Stress Mediated by Cocoa Polyphenols Extract. Molecules, 25(3), 452. https://doi.org/10.3390/molecules25030452