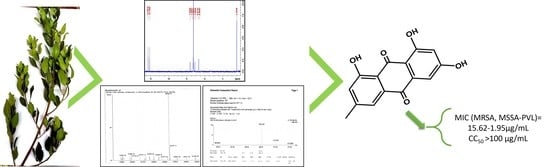
Bioguided Isolation of Active Compounds from Rhamnus alaternus against Methicillin-Resistant Staphylococcus aureus (MRSA) and Panton-Valentine Leucocidin Positive Strains (MSSA-PVL)
Abstract
:1. Introduction
2. Materials and Methods
2.1. General Procedure
2.2. Plant Material and Extraction
2.3. Bacterial Strains
2.4. Antibacterial Testing
2.4.1. Inoculum’s Preparation
2.4.2. Agar Well Diffusion Assay
2.4.3. Agar Dilution Assay
2.5. Bioguided Fractionation and Purification of Bioactive Compounds from the Leaves Extract of R. alaternus
2.5.1. Bioautography
2.5.2. Separation and Isolation of Pure Compounds
2.5.3. Chemical Structures Characterization
2.6. MIC Determination of Fractions and Pure Compounds
2.7. Cytotoxicity Assay
3. Results
3.1. Preliminary Antistaphylococcal Screening
3.2. MIC Determination
3.3. Bioguided Fractionation and Identification of Active Compounds
3.4. Cytotoxicity Assay
4. Discussion
5. Conclusions
6. Patent
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- WHO. The World Health Report 1996—Fighting Disease, Fostering Development; WHO: Geneva, Switzerland, 1997; Volume 18. [Google Scholar]
- Ebani, V.V. Biology and pathogenesis of Staphylococcus infection. Microorganisms 2020, 8, 383. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Duployez, C.; Guern, R.L.; Tinez, C.; Lejeune, A.; Robriquet, L.; Six, S.; Loïez, C.; Wallet, F. Panton–Valentine Leukocidin–Secreting. Emerg. Infect. Dis. 2020, 26, 1939–1941. [Google Scholar] [CrossRef]
- Nakaminami, H.; Ozawa, K.; Sasai, N.; Ikeda, M.; Nemoto, O.; Baba, N.; Matsuzaki, Y.; Sawamura, D.; Shimoe, F.; Inaba, Y.; et al. Current status of Panton–Valentine Leukocidin—Positive methicillin-resistant Staphylococcus aureus isolated from patients with skin and soft tissue infections in Japan. J. Dermatol. 2020, 47, 1280–1286. [Google Scholar] [CrossRef] [PubMed]
- Petraitiene, B.; Conejo, P.R.; Jankauskaite, L.; Kevalas, R.; Trumpulyte, G.; Snipaitiene, A.; Vitkauskiene, A.; Gurskis, V. Prevalence, clinical expression, invasiveness and outcome of Staphylococcus aureus containing Panton-Valentine leukocidin in children treated in a university hospital of Lithuania. Infect. Dis. 2020, 52, 464–472. [Google Scholar] [CrossRef] [PubMed]
- Chow, A.; Htun, H.L.; Hon, P.Y.; Ang, B.; Kanagasabai, K.; Koh, J.; Holden, M.T.G.; Hsu, L.Y. Comparative epidemiology and factors associated with major healthcare-associated methicillin-resistant Staphylococcus aureus clones among interconnected acute-, intermediate- and long-term healthcare facilities in Singapore. Clin. Microbiol. Infect. 2020. [Google Scholar] [CrossRef]
- Grousd, J.A.; Rich, H.E.; Alcorn, J.F. Host—Pathogen interactions in gram-positive bacterial pneumonia. Clin. Microbiol. Rev. 2019, 32, 1–22. [Google Scholar] [CrossRef]
- Aguilar, J.; Urday-Cornejo, V.; Donabedian, S.; Perri, M.; Tibbetts, R.; Zervos, M. Staphylococcus aureus meningitis case series and literature review. Medicine 2010, 89, 117–125. [Google Scholar] [CrossRef] [PubMed]
- Bawazir, Y.M.; Mustafa, M.A. Acute Esophageal Necrosis Associated With Methicillin—Resistant Staphylococcus Aureus Septicemia: A Case Report. Cureus 2020, 12. [Google Scholar] [CrossRef]
- Weiss, L.; Lansell, A.; Figueroa, J.; Suchdev, P.S.; Kirpalani, A. Declining prevalence of methicillin-resistant Staphylococcus aureus septic arthritis and osteomyelitis in children: Implications for treatment. Antibiotics 2020, 9, 101. [Google Scholar] [CrossRef] [PubMed]
- Selton-Suty, C.; Célard, M.; Le Moing, V.; Doco-Lecompte, T.; Chirouze, C.; Iung, B.; Strady, C.; Revest, M.; Vandenesch, F.; Bouvet, A.; et al. Preeminence of staphylococcus aureus in infective endocarditis: A 1-year population-based survey. Clin. Infect. Dis. 2012, 54, 1230–1239. [Google Scholar] [CrossRef] [Green Version]
- Brickner, S.J.; Barbachyn, M.R.; Hutchinson, D.K.; Manninen, P.R. Linezolid (ZYVOX), the first member of a completely new class of antibacterial agents for treatment of serious Gram-positive infections. J. Med. Chem. 2008, 51, 1981–1990. [Google Scholar] [CrossRef]
- Tsiodras, S.; Gold, H.S.; Sakoulas, G.; Eliopoulos, G.M.; Wennersten, C.; Venkataraman, L.; Moellering, R.C.; Ferraro, M.J. Linezolid resistance in a clinical isolate of Staphylococcus aureus. Lancet 2001, 358, 207–208. [Google Scholar] [CrossRef]
- Rouard, C.; Garnier, F.; Leraut, J.; Lepainteur, M.; Rahajamananav, L.; Languepin, J.; Ploy, M.C.; Bourgeois-Nicolaos, N.; Doucet-Populaire, F. Emergence and within-host genetic evolution of methicillin-resistant staphylococcus aureus resistant to linezolid in a cystic fibrosis patient. Antimicrob. Agents Chemother. 2018, 62, 1–11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ammar, R.B.; Kilani, S.; Bouhlel, I.; Ezzi, L.; Skandrani, I.; Boubaker, J.; Sghaier, M.B.; Naffeti, A.; Mahmoud, A.; Chekir-Ghedira, L.; et al. Antiproliferative, antioxidant, and antimutagenic activities of flavonoid-enriched extracts from (Tunisian) Rhamnus alaternus L.: Combination with the phytochemical composition. Drug Chem. Toxicol. 2008, 31, 61–80. [Google Scholar] [CrossRef] [PubMed]
- Cuoco, G.; Mathe, C.; Vieillescazes, C. Liquid chromatographic analysis of flavonol compounds in green fruits of three Rhamnus species used in Stil de grain. Microchem. J. 2014, 115, 130–137. [Google Scholar] [CrossRef]
- Longo, L.; Vasapollo, G.; Rescio, L. Identification of anthocyanins in Rhamnus alaternus L. berries. J. Agric. Food Chem. 2005, 53, 1723–1727. [Google Scholar] [CrossRef] [PubMed]
- Ammar, R.B.; Miyamoto, T.; Chekir-Ghedira, L.; Ghedira, K.; Lacaille-Dubois, M.A. Isolation and identification of new anthraquinones from Rhamnus alaternus L and evaluation of their free radical scavenging activity. Nat. Prod. Res. 2019, 33, 280–286. [Google Scholar] [CrossRef]
- Izhaki, I.; Tsahar, E.; Paluy, I.; Friedman, J. Within population variation and interrelationships between morphology, nutritional content, and secondary compounds of Rhamnus alaternus fruits. New Phytol. 2002, 156, 217–223. [Google Scholar] [CrossRef] [Green Version]
- Bhouri, W.; Sghaier, M.B.; Kilani, S.; Bouhlel, I.; Dijoux-Franca, M.G.; Ghedira, K.; Ghedira, L.C. Evaluation of antioxidant and antigenotoxic activity of two flavonoids from Rhamnus alaternus L. (Rhamnaceae): Kaempferol 3-O-β-isorhamninoside and rhamnocitrin 3-O-β-isorhamninoside. Food Chem. Toxicol. 2011, 49, 1167–1173. [Google Scholar] [CrossRef] [PubMed]
- Tacherfiout, M.; Petrov, P.D.; Mattonai, M.; Ribechini, E.; Ribot, J.; Bonet, M.L.; Khettal, B. Antihyperlipidemic effect of a Rhamnus alaternus leaf extract in Triton-induced hyperlipidemic rats and human HepG2 cells. Biomed. Pharmacother. 2018, 101, 501–509. [Google Scholar] [CrossRef]
- Ammar, R.B.; Bhouri, W.; Sghaier, M.B.; Boubaker, J.; Skandrani, I.; Neffati, A.; Bouhlel, I.; Kilani, S.; Mariotte, A.M.; Chekir-Ghedira, L.; et al. Antioxidant and free radical-scavenging properties of three flavonoids isolated from the leaves of Rhamnus alaternus L. (Rhamnaceae): A structure-activity relationship study. Food Chem. 2009, 116, 258–264. [Google Scholar] [CrossRef]
- Balouiri, M.; Sadiki, M.; Ibnsouda, S.K. Methods for in vitro evaluating antimicrobial activity: A review. J. Pharm. Anal. 2016, 6, 71–79. [Google Scholar] [CrossRef] [Green Version]
- Dewanjee, S.; Gangopadhyay, M.; Bhattacharya, N.; Khanra, R.; Dua, T.K. Bioautography and its scope in the field of natural product chemistry. J. Pharm. Anal. 2015, 5, 75–84. [Google Scholar] [CrossRef] [Green Version]
- Annuryanti, F.; Isnaeni, I.; Darmawati, A.; Rosyidah, I.; Dwiana, A.N. Method validation of contact and immersion tlc-bioautography for determination of streptomycin sulfate in shrimp. Turk. J. Pharm. Sci. 2020, 17, 254–258. [Google Scholar] [CrossRef] [PubMed]
- Zeouk, I.; Ouedrhiri, W.; Jiménez, I.A.; Morales, J.L.; Bazzocchi, I.L.; Bekhti, K. Intra-combined antioxidant activity and chemical characterization of three fractions from Rhamnus alaternus extract: Mixture design. Ind. Crop. Prod. 2020, 144, 112054. [Google Scholar] [CrossRef]
- Sifaoui, I.; López-Arencibia, A.; Martín-Navarro, C.M.; Reyes-Batlle, M.; Wagner, C.; Chiboub, O.; Mejri, M.; Valladares, B.; Abderrabba, M.; Piñero, J.E.; et al. Programmed cell death in Acanthamoeba castellanii Neff induced by several molecules present in olive leaf extracts. PLoS ONE 2017, 12, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Zeouk, I.; Ouali Lalami, A.E.; Bekhti, K. In Vitro Antibacterial Activity of Medicinal Plants in the Central North of Morocco: A Possible Source of Alternative Drugs Against Methicillin—Resistant Staphylococcus Aureus. Asian J. Pharm. Clin. Res. 2019, 12, 285–292. [Google Scholar] [CrossRef]
- Mari, S.H.; Varras, P.C.; Atia-Tul-Wahab; Choudhary, I.M.; Siskos, M.G.; Gerothanassis, I.P. Solvent—Dependent structures of natural products based on the combined use of DFT calculations and 1H-NMR chemical shifts. Molecules 2019, 24, 2290. [Google Scholar] [CrossRef] [Green Version]
- Demmak, R.G.; Bordage, S.; Bensegueni, A.; Boutaghane, N.; Hennebelle, T.; Mokrani, E.H.; Sahpaz, S. Chemical constituents from solenostemma argel and their cholinesterase inhibitory activity. Nat. Prod. Sci. 2019, 25, 115–121. [Google Scholar] [CrossRef] [Green Version]
- Guo, Y.; Song, G.; Sun, M.; Wang, J.; Wang, Y. Prevalence and Therapies of Antibiotic-Resistance in Staphylococcus aureus. Front. Cell. Infect. Microbiol. 2020, 10, 1–11. [Google Scholar] [CrossRef] [Green Version]
- Scudiero, O.; Brancaccio, M.; Mennitti, C.; Laneri, S.; Lombardo, B.; De Biasi, M.G.; De Gregorio, E.; Pagliuca, C.; Colicchio, R.; Salvatore, P.; et al. Human defensins: A novel approach in the fight against skin colonizing staphylococcus aureus. Antibiotics 2020, 9, 198. [Google Scholar] [CrossRef] [Green Version]
- David, M.Z.; Daum, R.S. Community—Associated methicillin-resistant Staphylococcus aureus: Epidemiology and clinical consequences of an emerging epidemic. Clin. Microbiol. Rev. 2010, 23, 616–687. [Google Scholar] [CrossRef] [Green Version]
- Gill, V.C.; Ma, I.; Guo, M.; Gregson, D.B.; Naugler, C.; Church, D.L. Sociodemographic and geospatial associations with community-associated methicillin-resistant Staphylococcus aureus (CA-MRSA) infections in a large Canadian city: An 11 year retrospective study. BMC Public Health 2019, 19, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Towers, S.; Panchanathan, S.; Chowell, G. A Population Based Study of Seasonality of Skin and Soft Tissue Infections: Implications for the Spread of CA-MRSA. PLoS ONE 2013, 8. [Google Scholar] [CrossRef]
- Ammar, R.B.; Kilani, S.; Bouhlel, I.; Skandrani, I.; Naffeti, A.; Boubaker, J.; Ben Sghaier, M.; Bhouri, W.; Mahmoud, A.; Chekir-Ghedira, L.; et al. Antibacterial and cytotoxic activities of extracts from (Tunisian) Rhamnus alaternus (Rhamnaceae). Ann. Microbiol. 2007, 57, 453–460. [Google Scholar] [CrossRef]
- Kosalec, I.; Kremer, D.; Locatelli, M.; Epifano, F.; Genovese, S.; Carlucci, G.; Randić, M.; Zovko Končić, M. Anthraquinone profile, antioxidant and antimicrobial activity of bark extracts of Rhamnus alaternus, R. fallax, R. intermedia and R. pumila. Food Chem. 2013, 136, 335–341. [Google Scholar] [CrossRef]
- Dong, X.; Fu, J.; Yin, X.; Cao, S.; Li, X.; Lin, L.; Huyiligequi; Ni, J. Emodin: A review of its pharmacology, toxicity and pharmacokinetics. Phyther. Res. 2016, 30, 1207–1218. [Google Scholar] [CrossRef] [PubMed]
- Basu, S.; Ghosh, A.; Hazra, B. Evaluation of the antibacterial activity of Ventilago madraspatana Gaertn., Rubia cordifolia Linn., and Lantana camara Linn.: Isolation of emodin and physcion as active antibacterial agents. Phyther. Res. 2005, 19, 888–894. [Google Scholar] [CrossRef] [PubMed]
- Chukwujekwu, J.C.; Coombes, P.H.; Mulholland, D.A.; van Staden, J. Emodin, an antibacterial anthraquinone from the roots of Cassia occidentalis. South Afr. J. Bot. 2006, 72, 295–297. [Google Scholar] [CrossRef] [Green Version]
- Yanwen, W.; Wenyuan, G.; Xiaohe, X.; Yi, L. Calorimetric investigation of the effect of hydroxyanthraquinones in Rheum officinale Baill on Staphylococcus aureus growth. Thermochim. Acta 2005, 429, 167–170. [Google Scholar] [CrossRef]
- Cao, F.; Peng, W.; Li, X.; Liu, M.; Li, B.; Qin, R.; Jiang, W.; Cen, Y.; Pan, X.; Yan, Z.; et al. Emodin is identified as the active component of ether extracts from Rhizoma Polygoni Cuspidati, for anti-MRSA activity. Can. J. Physiol. Pharmacol. 2015, 93, 1–9. [Google Scholar] [CrossRef]
- Ji, X.; Liu, X.; Peng, Y.; Zhan, R.; Xu, H.; Ge, X. Comparative analysis of methicillin-sensitive and resistant Staphylococcus aureus exposed to emodin based on proteomic profiling. Biochem. Biophys. Res. Commun. 2017, 494, 318–324. [Google Scholar] [CrossRef]
- Liu, B.; Yuan, B.; Zhang, L.; Mu, W.; Wang, C. ROS/p38/p53/Puma signaling pathway is involved in emodin-induced apoptosis of human colorectal cancer cells. Int. J. Clin. Exp. Med. 2015, 8, 15413–15422. [Google Scholar]
- Yan, X.; Gu, S.; Shi, Y.; Cui, X.; Wen, S.; Ge, J. The effect of emodin on Staphylococcus aureus strains in planktonic form and biofilm formation in vitro. Arch. Microbiol. 2017, 199, 1267–1275. [Google Scholar] [CrossRef] [PubMed]
- Hou, X.; Wei, W.; Fan, Y.; Zhang, J.; Zhu, N.; Hong, H.; Wang, C. Study on synthesis and bioactivity of biotinylated emodin. Appl. Microbiol. Biotechnol. 2017, 101, 5259–5266. [Google Scholar] [CrossRef] [PubMed]
- Chalothorn, T.; Rukachaisirikul, V.; Phongpaichit, S.; Pannara, S.; Tansakul, C. Synthesis and antibacterial activity of emodin and its derivatives against methicillin-resistant Staphylococcus aureus. Tetrahedron Lett. 2019, 60, 151004. [Google Scholar] [CrossRef]
- Kemegne, G.A.; Mkounga, P.; Essia Ngang, J.J.; Sado Kamdem, S.L.; Nkengfack, A.E. Antimicrobial structure activity relationship of five anthraquinones of emodine type isolated from Vismia laurentii. BMC Microbiol. 2017, 17, 1–8. [Google Scholar] [CrossRef] [Green Version]
- Lee, Y.S.; Kang, O.H.; Choi, J.G.; Oh, Y.C.; Keum, J.H.; Kim, S.B.; Jeong, G.S.; Kim, Y.C.; Shin, D.W.; Kwon, D.Y. Synergistic effect of emodin in combination with ampicillin or oxacillin against methicillin-resistant Staphylococcus aureus. Pharm. Biol. 2010, 48, 1285–1290. [Google Scholar] [CrossRef] [PubMed]
- Shi, Y.; Li, H.; Li, J.; Zhi, D.; Zhang, X.; Liu, H.; Wang, H.; Li, H. Development, optimization and evaluation of emodin loaded nanoemulsion prepared by ultrasonic emulsification. J. Drug Deliv. Sci. Technol. 2015, 27, 46–55. [Google Scholar] [CrossRef]
- Di, X.; Wang, X.; Liu, Y. Effect of piperine on the bioavailability and pharmacokinetics of emodin in rats. J. Pharm. Biomed. Anal. 2015, 115, 144–149. [Google Scholar] [CrossRef]
- Li, H.; Yang, T.; Zhou, H.; Du, J.; Zhu, B.; Sun, Z. Emodin combined with nanosilver inhibited sepsis by anti-inflammatory protection. Front. Pharmacol. 2017, 7. [Google Scholar] [CrossRef] [Green Version]
- Ban, E.; An, S.H.; Park, B.; Park, M.; Yoon, N.E.; Jung, B.H.; Kim, A. Improved Solubility and Oral Absorption of Emodin-Nicotinamide Cocrystal Over Emodin with PVP as a Solubility Enhancer and Crystallization Inhibitor. J. Pharm. Sci. 2020, 109, 3660–3667. [Google Scholar] [CrossRef]
- Wang, D.; Sun, M.; Zhang, Y.; Chen, Z.; Zang, S.; Li, G.; Li, G.; Clark, A.R.; Huang, J.; Si, L. Enhanced therapeutic efficacy of a novel colon-specific nanosystem loading emodin on DSS-induced experimental colitis. Phytomedicine 2020, 78, 153293. [Google Scholar] [CrossRef]
- Falcão-Silva, V.S.; Silva, D.A.; Souza, M.d.F.V.; Siqueira-Junior, J.P. Modulation of Drug Resistance in Staphylococcus aureus by a Kaempferol Glycoside from Herissantia tiubae (Malvaceae) 1367. Phyther. Res. 2009, 23, 1367–1370. [Google Scholar] [CrossRef] [PubMed]
- Ding, S.T.; He, X.P.; Ma, X.J.; Zhang, Y.; Liu, X.X.; Qin, J. Lactational Breast Abscesses Caused by Methicillin-Resistant or Methicillin-Sensitive Staphylococcus aureus Infection and Therapeutic Effect of Ultrasound-Guided Aspiration. Breastfeed. Med. 2020, 15, 471–474. [Google Scholar] [CrossRef]
- Crandall, H.; Kapusta, A.; Killpack, J.; Heyrend, C.; Nilsson, K.; Dickey, M.; Daly, J.A.; Ampofo, K.; Pavia, A.T.; Mulvey, M.A.; et al. Clinical and molecular epidemiology of invasive Staphylococcus aureus infection in Utah children; continued dominance of MSSA over MRSA. PLoS ONE 2020, 15, 1–14. [Google Scholar] [CrossRef]
- Sapri, H.F.; Sani, N.A.M.; Neoh, H.-M.; Hussin, S. Epidemiological Study on Staphylococcus aureus Isolates Reveals Inverse Relationship between Antibiotic Resistance and Virulence Repertoire. Indian J. Microbiol. 2013, 53, 321–322. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cameron, D.R.; Howden, B.P.; Peleg, A.Y. The interface between antibiotic resistance and virulence in staphylococcus aureus and its impact upon clinical outcomes. Clin. Infect. Dis. 2011, 53, 576–582. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schroeder, M.; Brooks, B.D.; Brooks, A.E. The complex relationship between virulence and antibiotic resistance. Genes 2017, 8, 39. [Google Scholar] [CrossRef] [PubMed] [Green Version]


| Ethanolic Extracts | S. aureus Strains | |||
|---|---|---|---|---|
| SaPVL+ | MRSA348 | Sa | MRSA (ATCC29213) | |
| R. alaternus | 14 ± 1.0 | 15 ± 1.0 | 25.5 ± 1.5 | 25 ± 1.0 |
| I. viscosa | 12 ± 1.0 | 12 ± 0.81 | 13.66 ± 1.88 | 13 ± 0.81 |
| C. oxyacantha | 11± 0.5 | 14 ± 0.5 | 12.33 ± 1.88 | 11.66 ± 1.24 |
| R. tinctorum | 10 ± 0.5 | 20 ± 0.5 | 10.33 ± 0.47 | 12.5 ± 0.5 |
| A. herba alba | 13 ± 1.88 | 10 ± 0.0 | 14.5 ± 2.12 | 13 ± 0.7 |
| B. hispanica | 15 ± 0.0 | 20 ± 1.0 | 26 ± 1.0 | 24 ± 1.0 |
| E. altissima | NA | NA | NA | NA |
| L. dentata | NA | NA | NA | NA |
| C. salviifolius | NA | NA | NA | NA |
| N. oleander | NA | NA | NA | NA |
| A. tinctoria | NA | NA | NA | NA |
| J. oxycedrus | NA | NA | NA | NA |
| U. dioica | NA | NA | NA | NA |
| G. alypum | NA | NA | NA | NA |
| A. majus | NA | NA | NA | NA |
| Ethanolic Extracts | MIC (mg/mL) | |||
|---|---|---|---|---|
| SaPVL+ | MRSA348 | Sa | MRSA (ATCC 29213) | |
| A. herba alba | >16 | 16 | 8 | 8 |
| B. hispanica | >16 | 16 | 16 | 16 |
| C. oxyacantha | 16 | 16 | 16 | 16 |
| I. viscosa | 8 | 4 | 4 | 4 |
| R. alaternus | 1.0 | 1.0 | 0.5 | 0.5 |
| R. tinctorum | >16 | 8 | 16 | 16 |
| Strains | SaPVL+ | MRSA348 | Sa | MRSA (ATCC 29213) |
|---|---|---|---|---|
| R. alaternus | MIC (µg/mL) | |||
| Crude extract | 1000 | 1000 | 500 | 500 |
| F1-F2 | NA | NA | NA | NA |
| F3 | 125 | 125 | 31,5 | 125 |
| F4 | NA | NA | NA | NA |
| F5 | DN | DN | DN | DN |
| F6 | 50 | 50 | 200 | 50 |
| F7-F12 | >1000 | >1000 | >1000 | >1000 |
| Control + | NG | NG | NG | NG |
| Control − | NG | NG | NG | NG |
| Compound/Strain | SaPVL+ | MRSA348 | Sa | MRSA (ATCC 29213) | Murin Macrophages | ||||
|---|---|---|---|---|---|---|---|---|---|
| MIC (µg/mL) | SI | MIC (µg/mL) | SI | MIC (µg/mL) | SI | MIC (µg/mL) | SI | CC50 (µg/mL) | |
| Emodin | 15.63 | >6.4 | 15.63 | >6.4 | 1.95 | >51.28 | 15.63 | >6.4 | >100 |
| Kaempferol | >250 | - | >250 | - | >250 | - | >250 | - | - |
| Control + | NG | NG | NG | NG | |||||
| Control − | NG | NG | NG | NG | |||||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zeouk, I.; Ouedrhiri, W.; Sifaoui, I.; Bazzocchi, I.L.; Piñero, J.E.; Jiménez, I.A.; Lorenzo-Morales, J.; Bekhti, K. Bioguided Isolation of Active Compounds from Rhamnus alaternus against Methicillin-Resistant Staphylococcus aureus (MRSA) and Panton-Valentine Leucocidin Positive Strains (MSSA-PVL). Molecules 2021, 26, 4352. https://doi.org/10.3390/molecules26144352
Zeouk I, Ouedrhiri W, Sifaoui I, Bazzocchi IL, Piñero JE, Jiménez IA, Lorenzo-Morales J, Bekhti K. Bioguided Isolation of Active Compounds from Rhamnus alaternus against Methicillin-Resistant Staphylococcus aureus (MRSA) and Panton-Valentine Leucocidin Positive Strains (MSSA-PVL). Molecules. 2021; 26(14):4352. https://doi.org/10.3390/molecules26144352
Chicago/Turabian StyleZeouk, Ikrame, Wessal Ouedrhiri, Ines Sifaoui, Isabel L. Bazzocchi, José E. Piñero, Ignacio A. Jiménez, Jacob Lorenzo-Morales, and Khadija Bekhti. 2021. "Bioguided Isolation of Active Compounds from Rhamnus alaternus against Methicillin-Resistant Staphylococcus aureus (MRSA) and Panton-Valentine Leucocidin Positive Strains (MSSA-PVL)" Molecules 26, no. 14: 4352. https://doi.org/10.3390/molecules26144352
APA StyleZeouk, I., Ouedrhiri, W., Sifaoui, I., Bazzocchi, I. L., Piñero, J. E., Jiménez, I. A., Lorenzo-Morales, J., & Bekhti, K. (2021). Bioguided Isolation of Active Compounds from Rhamnus alaternus against Methicillin-Resistant Staphylococcus aureus (MRSA) and Panton-Valentine Leucocidin Positive Strains (MSSA-PVL). Molecules, 26(14), 4352. https://doi.org/10.3390/molecules26144352