Influence of Open Chain and Cyclic Structure of Peptidomimetics on Antibacterial Activity in E. coli Strains
Abstract
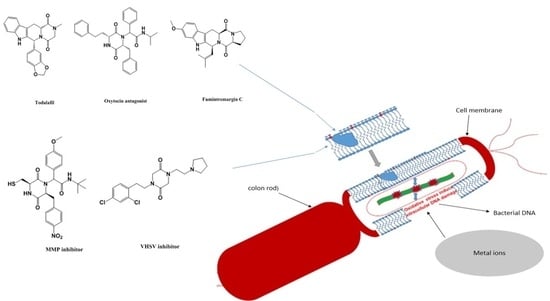
:1. Introduction
2. Materials and Methods
2.1. Microorganisms and Media
2.2. Chemicals
2.3. General Procedure for Synthesis of Compounds Va-Vh
2.3.1. N-benzyl-2-chloro-N-(2-((4-methoxybenzyl)amino)-2-oxo-1-phenylethyl) Propanamide (Va)
2.3.2. N-benzyl-2-chloro-N-(2-((4-methoxybenzyl)amino)-1-(4-methoxyphenyl)-2-oxoethyl) Propanamide (Vb)
2.3.3. N-benzyl-2-chloro-N-(2-(cyclohexylamino)-2-oxo-1-phenylethyl) Propanamide (Vc)
2.3.4. N-benzyl-2-chloro-N-(2-((4-methoxybenzyl)amino)-2-oxo-1-(p-tolyl)ethyl) Propanamide (Vd)
2.3.5. N-benzyl-2-chloro-N-(2-(cyclohexylamino)-2-oxo-1-(p-tolyl)ethyl) Propanamide (Ve)
2.3.6. N-benzyl-2-chloro-N-(2-(cyclohexylamino)-1-(2,4-dinitrophenyl)-2-oxoethyl) Propanamide (If)
2.3.7. N-benzyl-2-chloro-N-(2-(cyclohexylamino)-1-(4-(dimethylamino)phenyl)-2-oxoethyl) Propanamide (Vg)
2.3.8. N-benzyl-2-chloro-N-(2-(cyclohexylamino)-1-(4-nitrophenyl)-2-oxoethyl) Propanamide (Vh)
2.4. General Procedure for Synthesis of Compounds VIa-VIh
2.4.1. 1-benzyl-4-(4-methoxybenzyl)-3-methyl-6-phenylpiperazine-2,5-dione (VIa)
2.4.2. 1-benzyl-4-(4-methoxybenzyl)-6-(4-methoxyphenyl)-3-methylpiperazine-2,5-dione (VIb)
2.4.3. 1-benzyl-4-cyclohexyl-3-methyl-6-phenylpiperazine-2,5-dione (VIc)
2.4.4. 1-benzyl-4-(4-methoxybenzyl)-3-methyl-6-(p-tolyl)piperazine-2,5-dione (VId)
2.4.5. 1-benzyl-4-cyclohexyl-3-methyl-6-(p-tolyl)piperazine-2,5-dione (VIe)
2.4.6. 1-benzyl-4-cyclohexyl-6-(2,4-dinitrophenyl)-3-methylpiperazine-2,5-dione (VIf)
2.4.7. 1-benzyl-4-cyclohexyl-6-(4-(dimethylamino)phenyl)-3-methylpiperazine-2,5-dione (VIg)
2.4.8. 1-benzyl-4-cyclohexyl-3-methyl-6-(4-nitrophenyl)piperazine-2,5-dione (VIh)
3. Results & Discussion
3.1. Chemistry
3.2. Cytotoxic Studies of the Synthesized Compounds
3.3. Analysis of R2–R4 E. coli Strains Modified with Tested Compounds diketopiperazines
3.4. R2–R4 E. coli Strains with Tested Peptidomimetics
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
Abbreviations
| MIC | minimum inhibitory concentration |
| MBC | minimum bactericidal concentration |
| Oc | open circle |
| Ccc | covalently closed circle |
| BER | base excision repair |
| Fpg | DNA-formamidopyrimidine glycosylase |
References
- Ugi, I. The α-addition of immonium ions and anions to isonitriles accompanied by secondary reactions. Angew. Chem. Int. Ed. 1962, 1, 8–21. [Google Scholar] [CrossRef]
- Hulme, C.; Gore, V. Multi-component Reactions: Emerging Chemistry in Drug Discovery ‘From Xylocain to Crixivan’. Curr. Med. Chem. 2003, 10, 51–80. [Google Scholar] [CrossRef] [PubMed]
- Borthwick, A.D. 2, 5-Diketopiperazines: Synthesis, reactions, medicinal chemistry, and bioactive natural products. Chem. Rev. 2012, 112, 3641–3716. [Google Scholar] [CrossRef] [PubMed]
- Daugan, A.; Grondin, P.; Ruault, C.; Le Monnier de Gouville, A.-C.; Coste, H.; Linget, J.M.; Kirilovsky, J.; Hyafil, F.; Labaudiniere, R. The discovery of tadalafil: A novel and highly selective PDE5 inhibitor. 2: 2, 3, 6, 7, 12, 12a-hexahydropyrazino [1‘, 2‘: 1, 6] pyrido [3, 4-b] indole-1, 4-dione analogues. J. Med. Chem. 2003, 46, 4533–4542. [Google Scholar] [CrossRef] [PubMed]
- Wyatt, P.G.; Allen, M.J.; Borthwick, A.D.; Davies, D.E.; Exall, A.M.; Hatley, R.J.D.; Irving, W.R.; Livermore, D.G.; Miller, N.D.; Nerozzi, F.; et al. 2, 5-Diketopiperazines as potent and selective oxytocin antagonists 1: Identification, stereochemistry and initial SAR. Bioorg. Med. Chem. Lett. 2005, 15, 2579–2582. [Google Scholar] [CrossRef]
- Allen, J.D.; van Loevezijn, A.; Lakhai, J.M.; van der Valk, M.; van Tellingen, O.; Reid, G.; Schellens, J.H.M.; Koomen, G.J.; Schinkel, A.H. Potent and Specific Inhibition of the Breast Cancer Resistance Protein Multidrug Transporter in Vitro and in Mouse Intestine by a Novel Analogue of Fumitremorgin C1. Mol. Cancer Ther. 2002, 1, 417–425. [Google Scholar]
- Szardenings, A.K.; Antonenko, V.; Campbell, D.A.; DeFrancisco, N.; Ida, S.; Shi, L.H.; Sharkov, N.; Tien, D.; Wang, Y.W.; Navre, M. Identification of highly selective inhibitors of collagenase-1 from combinatorial libraries of diketopiperazines. J. Med. Chem. 1999, 42, 1348–1357. [Google Scholar] [CrossRef]
- Mas, V.; Falco, A.; Brocal, I.; Perez, L.; Coll, J.M.; Estepa, A. Identification of selective inhibitors of VHSV from biased combinatorial libraries of N, N′-disubstituted 2, 5-piperazinediones. Antivir. Res. 2006, 72, 107–115. [Google Scholar] [CrossRef]
- Miyoshi, T.; Miyairi, N.; Aoki, H.; Kohsaka, M.; Sakai, H.; Imanaka, H. Bicyclomycin, a new antibiotic I. Taxonomy, isolation and characterization. J. Antibiot. 1972, 25, 569–575. [Google Scholar] [CrossRef] [Green Version]
- Miyamura, S.; Ogasawara, N.; Otsuka, H.; Niwayama, S.; Tanaka, H.; Take, T.; Uchiyama, T.; Ochiai, H.; Abe, K. Antibiotic No. 5879, a new water-soluble antibiotic against Gram-negative bacteria. J. Antibiot. 1972, 25, 610–612. [Google Scholar] [CrossRef] [Green Version]
- Andersen, O.A.; Dixon, M.J.; Eggleston, I.M.; van Aalten, D.M. Natural product family 18 chitinase inhibitors. Nat. Prod. Rep. 2005, 22, 563–579. [Google Scholar] [CrossRef] [PubMed]
- Guan, J.; Mathai, S.; Harris, P.; Wen, J.Y.; Zhang, R.; Brimble, M.; Gluckman, P. Peripheral administration of a novel diketopiperazine, NNZ 2591, prevents brain injury and improves somatosensory-motor function following hypoxia–ischemia in adult rats. Neuropharmacology 2007, 53, 749–762. [Google Scholar] [CrossRef] [PubMed]
- Lopez-Rodrıguez, M.L.; Rosado, M.L.; Benhamu, B.; Morcillo, M.J.; Fernandez, E.; Schaper, K.J. Synthesis and Structure− Activity Relationships of a New Model of Arylpiperazines. 2. Three-Dimensional Quantitative Structure− Activity Relationships of Hydantoin−Phenylpiperazine Derivatives with Affinity for 5-HT1A and α1 Receptors. A Comparison of CoMFA Models. J. Med. Chem. 1997, 40, 1648–1656. [Google Scholar] [PubMed]
- Shimazaki, N.; Shima, I.; Okamoto, M.; Yoshida, K.; Hemmi, K.; Hashimoto, M. PAF inhibitory activity of diketopiperazines: Structure-activity relationships. In Platelet-Activating Factor and Structurally Related Alkyl Ehter Lipids. AOCS Pub. 1991, 26, 223–226. [Google Scholar]
- Bobylev, M.M.; Bobyleva, L.I.; Cutler, H.G.; Cutler, S.J.; Strobel, G.A. Growth regulating activity of maculosin analogs in the etiolated wheat coleoptile bioassay (Triticum aestivum L. cv. Wakeland). PGRSA Q. 1999, 27, 105–118. [Google Scholar]
- Williams, R.M.; Stocking, E.M.; Sanz-Cervera, J.F. Biosynthesis of prenylated alkaloids derived from tryptophan. Biosyn 2000, 209, 97–173. [Google Scholar]
- Kowalczyk, P.; Madej, A.; Szymczak, M.; Ostaszewski, R. α-Amidoamids as New Replacements of Antibiotics—Research on the Chosen K12, R2–R4 E. coli Strains. Materials 2020, 13, 5169. [Google Scholar] [CrossRef]
- Ku, I.W.; Kang, S.B.; Keum, G.C.; Kim, Y.S. Synthesis of α, β-unsaturated 2-Oxopyrrolidinyl acetamide derivatives by application of the Ugi/RCM reaction sequence. Bull. Korean Chem. Soc. 2011, 32, 3167–3170. [Google Scholar] [CrossRef] [Green Version]
- Sutanto, F.; Shaabani, S.; Oerlemans, R.; Eris, D.; Patil, P.; Hadian, M.; Dömling, A. Combining High-Throughput Synthesis and High-Throughput Protein Crystallography for Accelerated Hit Identification. Angew. Chem. Int. Ed. 2021, 60, 18231–18239. [Google Scholar] [CrossRef]
- Wang, W.; Wang, Y.; Tao, H.; Peng, X.; Liu, P.; Zhu, W. Cerebrosides of the halotolerant fungus Alternaria raphani isolated from a sea salt field. J. Nat. Prod. 2009, 72, 1695–1698. [Google Scholar] [CrossRef]
- Samsonowicz-Górski, J.; Kowalczyk, P.; Koszelewski, D.; Brodzka, A.; Szymczak, M.; Kramkowski, K.; Ostaszewski, R. The Synthesis and Evaluation of Amidoximes as Cytotoxic Agents on Model Bacterial E. coli Strains. Materials 2021, 14, 7577. [Google Scholar] [CrossRef] [PubMed]
- Kowalczyk, P.; Trzepizur, D.; Szymczak, M.; Skiba, G.; Kramkowski, K.; Ostaszewski, R. 1,2-Diarylethanols—A New Class of Compounds That Are Toxic to E. coli K12, R2–R4 Strains. Materials 2021, 14, 1025. [Google Scholar] [CrossRef] [PubMed]
- Laskowska, M.; Pastukh, O.; Fedorchuk, A.; Schabikowski, M.; Kowalczyk, P.; Zalasiński, M.; Laskowski, Ł. Nanostructured Silica with Anchoring Units: The 2D Solid Solvent for Molecules and Metal Ions. Int. J. Mol. Sci. 2020, 21, 8137. [Google Scholar] [CrossRef]
- Kowalczyk, P.; Borkowski, A.; Czerwonka, G.; Cłapa, T.; Cieśla, J.; Misiewicz, A.; Borowiec, M.; Szala, M. The microbial toxicity of quaternary ammonium ionic liquids is dependent on the type of lipopolysaccharide. J. Mol. Liq. 2018, 266, 540–547. [Google Scholar] [CrossRef]
- Borkowski, A.; Kowalczyk, P.; Czerwonka, G.; Ciésla, J.; Cłapa, T.; Misiewicz, A.; Szala, M.; Drabik, M. Interaction of quaternary ammonium ionic liquids with bacterial membranes—Studies with Escherichia coli R1–R4-type lipopolysaccharides. J. Mol. Liq. 2017, 246, 282–289. [Google Scholar] [CrossRef]
- Kowalczyk, P.; Gawdzik, B.; Trzepizur, D.; Szymczak, M.; Skiba, G.; Raj, S.; Kramkowski, K.; Lizut, R.; Ostaszewski, R. δ-Lactones—A New Class of Compounds That Are Toxic to E. coli K12 and R2–R4 Strains. Materials 2021, 14, 2956. [Google Scholar] [CrossRef]
- Maciejewska, A.; Kaszowska, M.; Jachymek, W.; Lugowski, C.; Lukasiewicz, J. Lipopolysaccharide-linked Enterobacterial Common Antigen (ECALPS) Occurs in Rough Strains of Escherichia coli R1, R2, and R4. Int. J. Mol. Sci. 2020, 21, 6038. [Google Scholar] [CrossRef]
- Prost, M.E.; Prost, R. Basic parameters of evaluation of the effectiveness of antibiotic therapy. Ophth. Ther. 2017, 4, 233–236. [Google Scholar]
- Al-Jaidi, B.A.; Telfah, S.T.; Bardaweel, S.K.; Deb, P.K.; Borah, P.; Venugopala, K.N.; Bataineh, Y.A.; Al Khames Aga, Q.A. Anticancer Activity and In Silico ADMET Properties of 2,4,5-Trisubstitutedthiazole Derivatives. Curr. Drug Metab. 2021, 22, 532–536. [Google Scholar] [CrossRef]
- Simic, M.; Petkovic, M.; Jovanovic, P.; Jovanovic, M.; Tasic, G.; Besu, I.; Zizak, Z.; Aleksic, I.; Nikodinovic-Runic, J.; Savic, V. Fragment-type 4-azolylcoumarin derivatives with anticancer properties. Arch. Pharm. 2021, 354, 2100238. [Google Scholar] [CrossRef] [PubMed]
- Balalaie, S.; Ramezani Kejani, R.; Ghabraie, E.; Darvish, F.; Rominger, F.; Hamdan, F.; Bijanzadeh, H.R. Diastereoselective synthesis of functionalized diketopiperazines through post-transformational reactions. J. Org. Chem. 2017, 82, 12141–12152. [Google Scholar] [CrossRef] [PubMed]
- Halimehjani, A.Z.; Sharifi, M. Synthesis of a novel category of Ugi adducts using succinic acid, succinic anhydride and maleic anhydride and their application in post-Ugi reactions for synthesis of functionalized piperazine 2, 5-diones. Tetrahedron 2017, 73, 5778–5783. [Google Scholar] [CrossRef]
- Kowalczyk, P.; Wilk, M.; Parul, P.; Szymczak, M.; Kramkowski, K.; Raj, S.; Skiba, G.; Sulejczak, D.; Kleczkowska, P.; Ostaszewski, R. The Synthesis and Evaluation of Aminocoumarin Peptidomimetics as Cytotoxic Agents on Model Bacterial E. coli Strains. Materials 2021, 14, 5725. [Google Scholar] [CrossRef] [PubMed]
- Thompson, M.J.; Chen, B. Ugi reactions with ammonia offer rapid access to a wide range of 5-aminothiazole and oxazole derivatives. J. Org. Chem. 2009, 74, 7084–7093. [Google Scholar] [CrossRef] [PubMed]
- Radadia, A.C.; Rajapara, J.G.; Naliapara, Y.T. An efficient protocol for the synthesis of N-fused 2, 5-diketopiperazine via base catalyzed Ugi-type MCR. Org. Chem. 2018, 2018, 174–183. [Google Scholar] [CrossRef] [Green Version]
- Pobłocki, K.; Jacewicz, D.; Walczak, J.; Gawdzik, B.; Kramkowski, K.; Drzeżdżon, J.; Kowalczyk, P. Preparation of Allyl Alcohol Oligomers Using Dipicolinate Oxovanadium(IV) Coordination Compound. Materials 2022, 15, 695. [Google Scholar] [CrossRef] [PubMed]
- Qian, Z.; Liu, T.; Liu, Y.Y.; Briesewitz, R.; Barrios, A.M.; Jhiang, S.M.; Pei, D. Efficient delivery of cyclic peptides into mammalian cells with short sequence motifs. ACS Chem. Biol. 2013, 8, 423–431. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cascales, L.; Henriques, S.T.; Kerr, M.C.; Huang, Y.H.; Sweet, M.J.; Daly, N.L.; Craik, D.J. Identification and characterization of a new family of cell-penetrating peptides: Cyclic cell-penetrating peptides. J. Biol. Chem. 2011, 286, 36932–36943. [Google Scholar] [CrossRef] [Green Version]
- Mandal, D.; Shirazi, A.N.; Parang, K. Cell-penetrating homochiral cyclic peptides as nuclear-targeting molecular transporters. Angew. Chem. Int. Ed. 2011, 50, 9633–9637. [Google Scholar] [CrossRef]
- Traboulsi, H.; Larkin, H.; Bonin, M.A.; Volkov, L.; Lavoie, C.L.; Marsault, E. Macrocyclic cell penetrating peptides: A study of structure-penetration properties. Bioconjugate Chem. 2015, 26, 405–411. [Google Scholar] [CrossRef]
- Feni, L.; Jütten, L.; Parente, S.; Piarulli, U.; Neundorf, I.; Diaz, D. Cell-penetrating peptides containing 2, 5-diketopiperazine (DKP) scaffolds as shuttles for anti-cancer drugs: Conformational studies and biological activity. Chem. Commun. 2022, 56, 5685–5688. [Google Scholar] [CrossRef]











| Entry | Solvent | Temperature (°C) | Yield (%) |
|---|---|---|---|
| 1 | Methanol | 25 | 30 |
| 2 | Methanol | 30 | 32 |
| 3 | Methanol | 40 | 33 |
| 4 | Methanol | 50 | 39 |
| 5 | Methanol | 60 | 39 |
| 6 | Ethanol | 50 | 33 |
| 7 | Isopropanol | 50 | 37 |
| 8 | TFE | 50 | 53 |
| 9 | TFE | 60 | 42 |
| Entry | Base | Solvent | Temperature | Yield (%) |
|---|---|---|---|---|
| 1 | K2CO3 | DMF | 100 | 25 |
| 2 | K2CO3 | DMF | 90 | 27 |
| 3 | K2CO3 | DMF | 110 | 26 |
| 4 | K2CO3 | Ethanol | 50 | 30 |
| 5 | K2CO3 | Ethanol | 65 | 39 |
| 6 | K2CO3 | Ethanol | 75 | 28 |
| 7 | K2CO3 | THF | 65 | 49 |
| 8 | K2CO3 | DMF | 65 | 30 |
| 9 | K2CO3 | DMSO | 65 | 32 |
| 10 | K2CO3 | Toluene | 65 | 29 |
| 11 | K2CO3 | Methanol | 65 | 34 |
| 12 | CsF | THF | 65 | 33 |
| 13 | NaHCO3 | THF | 65 | 36 |
| 14 | KOtBu | THF | 65 | 27 |
| 15 | KOH | THF | 65 | 19 |
| 16 | NaH | THF | 65 | 70 |
| No. of Samples | Va | Vb | Vc | Vd | Ve | Vf | Vg | Vh | Type of Test |
|---|---|---|---|---|---|---|---|---|---|
| K12 | ** | ** | ** | ** | * | * | * | ** | MIC |
| R2 | ** | ** | ** | ** | * | * | * | ** | MIC |
| R3 | ** | ** | ** | ** | * | * | * | ** | MIC |
| R4 | ** | ** | ** | ** | * | * | * | ** | MIC |
| K12 | * | * | ** | * | ** | * | * | ** | MBC |
| R2 | * | * | ** | * | ** | * | * | ** | MBC |
| R3 | * | * | ** | * | ** | * | * | ** | MBC |
| R4 | * | * | ** | * | ** | * | * | ** | MBC |
| K12 | ** | * | * | * | * | * | * | *** | MBC/MIC |
| R2 | ** | * | * | * | * | * | ** | *** | MBC/MIC |
| R3 | ** | * | * | * | * | * | ** | *** | MBC/MIC |
| R4 | ** | * | * | * | * | * | ** | *** | MBC/MIC |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sahrawat, P.; Kowalczyk, P.; Koszelewski, D.; Szymczak, M.; Kramkowski, K.; Wypych, A.; Ostaszewski, R. Influence of Open Chain and Cyclic Structure of Peptidomimetics on Antibacterial Activity in E. coli Strains. Molecules 2022, 27, 3633. https://doi.org/10.3390/molecules27113633
Sahrawat P, Kowalczyk P, Koszelewski D, Szymczak M, Kramkowski K, Wypych A, Ostaszewski R. Influence of Open Chain and Cyclic Structure of Peptidomimetics on Antibacterial Activity in E. coli Strains. Molecules. 2022; 27(11):3633. https://doi.org/10.3390/molecules27113633
Chicago/Turabian StyleSahrawat, Parul, Paweł Kowalczyk, Dominik Koszelewski, Mateusz Szymczak, Karol Kramkowski, Aleksandra Wypych, and Ryszard Ostaszewski. 2022. "Influence of Open Chain and Cyclic Structure of Peptidomimetics on Antibacterial Activity in E. coli Strains" Molecules 27, no. 11: 3633. https://doi.org/10.3390/molecules27113633
APA StyleSahrawat, P., Kowalczyk, P., Koszelewski, D., Szymczak, M., Kramkowski, K., Wypych, A., & Ostaszewski, R. (2022). Influence of Open Chain and Cyclic Structure of Peptidomimetics on Antibacterial Activity in E. coli Strains. Molecules, 27(11), 3633. https://doi.org/10.3390/molecules27113633