Pyrazolo[4,3-e]tetrazolo[1,5-b][1,2,4]triazine Sulfonamides as Novel Potential Anticancer Agents: Cytotoxic and Genotoxic Activities In Vitro
Abstract
:1. Introduction
2. Results
2.1. Synthesis of Novel 1,2,4-Triazine Derivatives
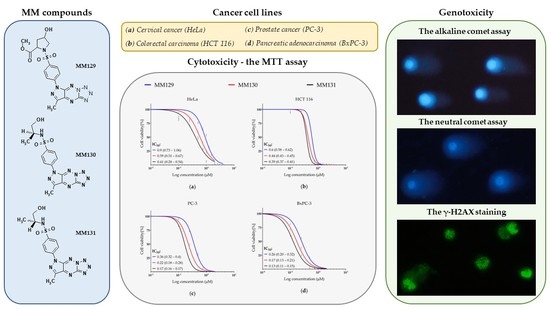
2.2. Cytotoxicity—MTT Assay
2.3. Genotoxicity
2.3.1. Comet Assay
Alkaline Version (pH > 13)
Neutral Version (pH 9.0)
2.3.2. Immunocytochemical Detection of γ-H2AX
3. Discussion
4. Materials and Methods
4.1. Synthesis
4.1.1. General
4.1.2. Preparation of N-(S)-(1-hydroxy-propan-2-yl)-4-(3-methyl-5-methylsulfonyl-1H-pyrazolo[4,3-e][1,2,4]triazin-1-yl)benzenesulfonamide (2b)
4.1.3. Synthesis of N-(S)-(1-hydroxy-propan-2-yl)-4-[7-methyl-5H-pyrazolo[4,3-e]tetrazolo[1,5-b][1,2,4]-triazin-5-yl)]benzenesulfonamide (MM130)
4.2. Chemicals
4.3. Cell Culture
4.4. Cytotoxicity—MTT Assay
4.5. Genotoxicity
4.5.1. Comet Assay
4.5.2. Immunocytochemical Detection of γ-H2AX
4.6. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Sample Availability
References
- Ferlay, J.; Colombet, M.; Soerjomataram, I.; Parkin, D.M.; Piñeros, M.; Znaor, A.; Bray, F. Cancer Statistics for the Year 2020: An Overview. Int. J. Cancer 2021, 149, 778–789. [Google Scholar] [CrossRef]
- World Health Organization. Cancer. Available online: https://www.who.int/news-room/fact-sheets/detail/cancer (accessed on 3 February 2022).
- Wang, X.; Zhang, H.; Chen, X. Drug Resistance and Combating Drug Resistance in Cancer. CDR 2019, 2, 141–160. [Google Scholar] [CrossRef] [PubMed]
- Majidinia, M.; Mirza-Aghazadeh-Attari, M.; Rahimi, M.; Mihanfar, A.; Karimian, A.; Safa, A.; Yousefi, B. Overcoming Multidrug Resistance in Cancer: Recent Progress in Nanotechnology and New Horizons. IUBMB Life 2020, 72, 855–871. [Google Scholar] [CrossRef] [PubMed]
- Nussinov, R.; Tsai, C.-J.; Jang, H. Anticancer Drug Resistance: An Update and Perspective. Drug Resist. Updates 2021, 59, 100796. [Google Scholar] [CrossRef]
- Bhimani, J.; Philipps, L.; Simpson, L.; Lythgoe, M.; Soultati, A.; Webb, A.; Savage, P. The Impact of New Cancer Drug Therapies on Site Specialised Cancer Treatment Activity in a UK Cancer Network 2014–2018. J. Oncol. Pharm. Pract. 2020, 26, 93–98. [Google Scholar] [CrossRef] [PubMed]
- Cross, D.; Burmester, J.K. Gene Therapy for Cancer Treatment: Past, Present and Future. Clin. Med. Res. 2006, 4, 218–227. [Google Scholar] [CrossRef]
- Montaño-Samaniego, M.; Bravo-Estupiñan, D.M.; Méndez-Guerrero, O.; Alarcón-Hernández, E.; Ibáñez-Hernández, M. Strategies for Targeting Gene Therapy in Cancer Cells With Tumor-Specific Promoters. Front. Oncol. 2020, 10, 605380. [Google Scholar] [CrossRef]
- Nussbaumer, S.; Bonnabry, P.; Veuthey, J.-L.; Fleury-Souverain, S. Analysis of Anticancer Drugs: A Review. Talanta 2011, 85, 2265–2289. [Google Scholar] [CrossRef]
- Marchi, E.; O’Connor, O.A. Safety and Efficacy of Pralatrexate in the Treatment of Patients with Relapsed or Refractory Peripheral T-Cell Lymphoma. Ther. Adv. Hematol. 2012, 3, 227–235. [Google Scholar] [CrossRef]
- Kabbaj, Y.; Lazrek, H.B.; Barascut, J.L.; Imbach, J.L. Synthesis and Biological Activity of Some Unsaturated 6-Azauracil Acyclonucleosides. Nucleosides Nucleotides Nucleic Acids 2005, 24, 161–172. [Google Scholar] [CrossRef]
- Saad, H.; Moustafa, A. Synthesis and Anticancer Activity of Some New S-Glycosyl and S-Alkyl 1,2,4-Triazinone Derivatives. Molecules 2011, 16, 5682–5700. [Google Scholar] [CrossRef] [PubMed]
- Kumar, R.; Sirohi, T.S.; Singh, H.; Yadav, R.; Roy, R.K.; Chaudhary, A.; Pandeya, S.N. 1,2,4-Triazine Analogs as Novel Class of Therapeutic Agents. MRMC 2014, 14, 168–207. [Google Scholar] [CrossRef] [PubMed]
- Cascioferro, S.; Parrino, B.; Spanò, V.; Carbone, A.; Montalbano, A.; Barraja, P.; Diana, P.; Cirrincione, G. An Overview on the Recent Developments of 1,2,4-Triazine Derivatives as Anticancer Compounds. Eur. J. Med. Chem. 2017, 142, 328–375. [Google Scholar] [CrossRef] [PubMed]
- Mojzych, M.; Rykowski, A.; Wierzchowski, J. Pyrazolo[4,3-e][1,2,4]Triazines: Purine Analogues with Electronic Absorption in the Visible Region. Molecules 2005, 10, 1298–1306. [Google Scholar] [CrossRef]
- Lindner, H.J.; Schaden, G. Pyrazolo[4.3-e]As-triazin, Ein Neues Heterocyclisches System Aus Pseudomonas Fluorescens Var. Pseudoiodinum. Chem. Ber. 1972, 105, 1949–1955. [Google Scholar] [CrossRef]
- Smirnov, V.V.; Kiprianova, E.A.; Garagulya, A.D.; Esipov, S.E.; Dovjenko, S.A. Fluviols, Bicyclic Nitrogen-Rich Antibiotics Produced by Pseudomonas Fluorescens. FEMS Microbiol. Lett. 1997, 153, 357–361. [Google Scholar] [CrossRef]
- Hirata, K.; Nakagami, H.; Takashina, J.; Mahmud, T.; Kobayashi, M.; In, Y.; Ishida, T.; Miyamoto, K. ChemInform Abstract: Novel Violet Pigment, Nostocine A, an Extracellular Metabolite from Cyanobacterium Nostoc Spongiaeforme. ChemInform 2010, 27, 1513–1519. [Google Scholar] [CrossRef]
- Mojzych, M.; Tarasiuk, P.; Kotwica-Mojzych, K.; Rafiq, M.; Seo, S.-Y.; Nicewicz, M.; Fornal, E. Synthesis of Chiral Pyrazolo[4,3-e][1,2,4]Triazine Sulfonamides with Tyrosinase and Urease Inhibitory Activity. J. Enzym. Inhib. Med. Chem. 2017, 32, 99–105. [Google Scholar] [CrossRef]
- Mojzych, M.; Bielawska, A.; Bielawski, K.; Ceruso, M.; Supuran, C.T. Pyrazolo[4,3-e][1,2,4]Triazine Sulfonamides as Carbonic Anhydrase Inhibitors with Antitumor Activity. Bioorg. Med. Chem. 2014, 22, 2643–2647. [Google Scholar] [CrossRef]
- Mojzych, M.; Ceruso, M.; Bielawska, A.; Bielawski, K.; Fornal, E.; Supuran, C.T. New Pyrazolo[4,3-e][1,2,4]Triazine Sulfonamides as Carbonic Anhydrase Inhibitors. Bioorg. Med. Chem. 2015, 23, 3674–3680. [Google Scholar] [CrossRef]
- Mojzych, M.; Šubertová, V.; Bielawska, A.; Bielawski, K.; Bazgier, V.; Berka, K.; Gucký, T.; Fornal, E.; Kryštof, V. Synthesis and Kinase Inhibitory Activity of New Sulfonamide Derivatives of Pyrazolo[4,3-e][1,2,4]Triazines. Eur. J. Med. Chem. 2014, 78, 217–224. [Google Scholar] [CrossRef]
- Manning, G.; Whyte, D.B.; Martinez, R.; Hunter, T.; Sudarsanam, S. The Protein Kinase Complement of the Human Genome. Science 2002, 298, 1912–1934. [Google Scholar] [CrossRef] [PubMed]
- Fabbro, D.; Cowan-Jacob, S.W.; Moebitz, H. Ten Things You Should Know about Protein Kinases: IUPHAR Review 14: Ten Things You Should Know about Protein Kinases. Br. J. Pharmacol. 2015, 172, 2675–2700. [Google Scholar] [CrossRef] [PubMed]
- Krystof, V.; Uldrijan, S. Cyclin-Dependent Kinase Inhibitors as Anticancer Drugs. CDT 2010, 11, 291–302. [Google Scholar] [CrossRef] [PubMed]
- Ades, F.; Metzger-Filho, O. Targeting the Cellular Signaling: BRAF Inhibition and Beyond for the Treatment of Metastatic Malignant Melanoma. Dermatol. Res. Pract. 2012, 2012, 259170. [Google Scholar] [CrossRef]
- Bedi, A.; Zehnbauer, B.; Barber, J.; Sharkis, S.; Jones, R. Inhibition of Apoptosis by BCR-ABL in Chronic Myeloid Leukemia. Blood 1994, 83, 2038–2044. [Google Scholar] [CrossRef]
- Bazzoni, G.; Carlesso, N.; Griffin, J.D.; Hemler, M.E. Bcr/Abl Expression Stimulates Integrin Function in Hematopoietic Cell Lines. J. Clin. Investig. 1996, 98, 521–528. [Google Scholar] [CrossRef]
- Cortez, D.; Stoica, G.; Pierce, J.H.; Pendergast, A.M. The BCR-ABL Tyrosine Kinase Inhibits Apoptosis by Activating a Ras-Dependent Signaling Pathway. Oncogene 1996, 13, 2589–2594. [Google Scholar]
- Cambier, N.; Chopra, R.; Strasser, A.; Metcalf, D.; Elefanty, A.G. BCR–ABL Activates Pathways Mediating Cytokine Independence and Protection against Apoptosis in Murine Hematopoietic Cells in a Dose-Dependent Manner. Oncogene 1998, 16, 335–348. [Google Scholar] [CrossRef]
- Carter, B.Z.; Mak, P.Y.; Mu, H.; Wang, X.; Tao, W.; Mak, D.H.; Dettman, E.J.; Cardone, M.; Zernovak, O.; Seki, T.; et al. Combined Inhibition of MDM2 and BCR-ABL1 Tyrosine Kinase Targets Chronic Myeloid Leukemia Stem/Progenitor Cells in a Murine Model. Haematologica 2020, 105, 1274–1284. [Google Scholar] [CrossRef]
- Bernat, Z.; Szymanowska, A.; Kciuk, M.; Kotwica-Mojzych, K.; Mojzych, M. Review of the Synthesis and Anticancer Properties of Pyrazolo[4,3-e][1,2,4]Triazine Derivatives. Molecules 2020, 25, 3948. [Google Scholar] [CrossRef] [PubMed]
- Ghorab, M.M.; Alsaid, M.S.; Abdullah-al-Dhfyan; Arafa, R.K. Cytotoxic Activity of Some Novel Sulfonamide Derivatives. Acta Pol. Pharm. 2015, 72, 79–87. [Google Scholar] [PubMed]
- Gornowicz, A.; Szymanowska, A.; Mojzych, M.; Bielawski, K.; Bielawska, A. The Effect of Novel 7-Methyl-5-Phenyl-Pyrazolo[4,3-e]Tetrazolo[4,5-b][1,2,4]Triazine Sulfonamide Derivatives on Apoptosis and Autophagy in DLD-1 and HT-29 Colon Cancer Cells. IJMS 2020, 21, 5221. [Google Scholar] [CrossRef] [PubMed]
- Hermanowicz, J.M.; Szymanowska, A.; Sieklucka, B.; Czarnomysy, R.; Pawlak, K.; Bielawska, A.; Bielawski, K.; Kalafut, J.; Przybyszewska, A.; Surazynski, A.; et al. Exploration of Novel Heterofused 1,2,4-Triazine Derivative in Colorectal Cancer. J. Enzym. Inhib. Med. Chem. 2021, 36, 535–548. [Google Scholar] [CrossRef] [PubMed]
- Hermanowicz, J.M.; Pawlak, K.; Sieklucka, B.; Czarnomysy, R.; Kwiatkowska, I.; Kazberuk, A.; Surazynski, A.; Mojzych, M.; Pawlak, D. MM-129 as a Novel Inhibitor Targeting PI3K/AKT/MTOR and PD-L1 in Colorectal Cancer. Cancers 2021, 13, 3203. [Google Scholar] [CrossRef]
- Gornowicz, A.; Szymanowska, A.; Mojzych, M.; Czarnomysy, R.; Bielawski, K.; Bielawska, A. The Anticancer Action of a Novel 1,2,4-Triazine Sulfonamide Derivative in Colon Cancer Cells. Molecules 2021, 26, 2045. [Google Scholar] [CrossRef]
- Adisty Ridha Damasuri; Eti Nurwening Sholikhah; Mustofa Cytotoxicity of ((E)-1-(4-Aminophenyl)-3-Phenylprop-2-En-1-One)) on HeLa Cell Line. Indones. J. Pharmacol. Ther. 2020, 1, 1–6. [CrossRef]
- Huanwen, W.; Zhiyong, L.; Xiaohua, S.; Xinyu, R.; Kai, W.; Tonghua, L. Intrinsic Chemoresistance to Gemcitabine Is Associated with Constitutive and Laminin-Induced Phosphorylation of FAK in Pancreatic Cancer Cell Lines. Mol. Cancer 2009, 8, 125. [Google Scholar] [CrossRef]
- Ikehata, M.; Ogawa, M.; Yamada, Y.; Tanaka, S.; Ueda, K.; Iwakawa, S. Different Effects of Epigenetic Modifiers on the Cytotoxicity Induced by 5-Fluorouracil, Irinotecan or Oxaliplatin in Colon Cancer Cells. Biol. Pharm. Bull. 2014, 37, 67–73. [Google Scholar] [CrossRef]
- Becit, M.; Aydın Dilsiz, S.; Başaran, N. Interaction of Curcumin on Cisplatin Cytotoxicity in HeLa and HepG2 Carcinoma Cells. Istanb. J. Pharm. 2020, 50, 202–210. [Google Scholar] [CrossRef]
- Barbanente, A.; Iacobazzi, R.M.; Azzariti, A.; Hoeschele, J.D.; Denora, N.; Papadia, P.; Pacifico, C.; Natile, G.; Margiotta, N. New Oxaliplatin-Pyrophosphato Analogs with Improved In Vitro Cytotoxicity. Molecules 2021, 26, 3417. [Google Scholar] [CrossRef] [PubMed]
- Aras, B.; Yerlikaya, A. Bortezomib and Etoposide Combinations Exert Synergistic Effects on the Human Prostate Cancer Cell Line PC-3. Oncol. Lett. 2016, 11, 3179–3184. [Google Scholar] [CrossRef] [PubMed]
- Fong, C.W. Platinum Anti-Cancer Drugs: Free Radical Mechanism of Pt-DNA Adduct Formation and Anti-Neoplastic Effect. Free Radic. Biol. Med. 2016, 95, 216–229. [Google Scholar] [CrossRef] [PubMed]
- Gajek, A.; Denel-Bobrowska, M.; Rogalska, A.; Bukowska, B.; Maszewski, J.; Marczak, A. Early Activation of Apoptosis and Caspase-Independent Cell Death Plays an Important Role in Mediating the Cytotoxic and Genotoxic Effects of WP 631 in Ovarian Cancer Cells. Asian Pac. J. Cancer Prev. 2016, 16, 8503–8512. [Google Scholar] [CrossRef]
- Stornetta, A.; Zimmermann, M.; Cimino, G.D.; Henderson, P.T.; Sturla, S.J. DNA Adducts from Anticancer Drugs as Candidate Predictive Markers for Precision Medicine. Chem. Res. Toxicol. 2017, 30, 388–409. [Google Scholar] [CrossRef]
- Burma, S.; Chen, B.P.; Murphy, M.; Kurimasa, A.; Chen, D.J. ATM Phosphorylates Histone H2AX in Response to DNA Double-Strand Breaks. J. Biol. Chem. 2001, 276, 42462–42467. [Google Scholar] [CrossRef]
- Rogakou, E.P.; Pilch, D.R.; Orr, A.H.; Ivanova, V.S.; Bonner, W.M. DNA Double-Stranded Breaks Induce Histone H2AX Phosphorylation on Serine 139. J. Biol. Chem. 1998, 273, 5858–5868. [Google Scholar] [CrossRef]
- Rogakou, E.P.; Boon, C.; Redon, C.; Bonner, W.M. Megabase Chromatin Domains Involved in DNA Double-Strand Breaks in Vivo. J. Cell Biol. 1999, 146, 905–916. [Google Scholar] [CrossRef]
- Rothkamm, K.; Lobrich, M. Evidence for a Lack of DNA Double-Strand Break Repair in Human Cells Exposed to Very Low x-Ray Doses. Proc. Natl. Acad. Sci. USA 2003, 100, 5057–5062. [Google Scholar] [CrossRef]
- Rothkamm, K.; Horn, S.; Scherthan, H.; Rößler, U.; De Amicis, A.; Barnard, S.; Kulka, U.; Lista, F.; Meineke, V.; Braselmann, H.; et al. Laboratory Intercomparison on the γ-H2AX Foci Assay. Radiat. Res. 2013, 180, 149. [Google Scholar] [CrossRef]
- Meyer, B.; Voss, K.-O.; Tobias, F.; Jakob, B.; Durante, M.; Taucher-Scholz, G. Clustered DNA Damage Induces Pan-Nuclear H2AX Phosphorylation Mediated by ATM and DNA–PK. Nucleic Acids Res. 2013, 41, 6109–6118. [Google Scholar] [CrossRef] [PubMed]
- Noubissi, F.K.; McBride, A.A.; Leppert, H.G.; Millet, L.J.; Wang, X.; Davern, S.M. Detection and Quantification of γ-H2AX Using a Dissociation Enhanced Lanthanide Fluorescence Immunoassay. Sci. Rep. 2021, 11, 8945. [Google Scholar] [CrossRef] [PubMed]
- de Lapuente, J.; Lourenço, J.; Mendo, S.A.; Borràs, M.; Martins, M.G.; Costa, P.M.; Pacheco, M. The Comet Assay and Its Applications in the Field of Ecotoxicology: A Mature Tool That Continues to Expand Its Perspectives. Front. Genet. 2015, 6, 180. [Google Scholar] [CrossRef] [PubMed]
- Chhabra, N.; Aseri, M.L.; Padmanabhan, D. A Review of Drug Isomerism and Its Significance. Int. J. Appl. Basic Med. Res. 2013, 3, 16–18. [Google Scholar] [CrossRef]
- ISO-10993-5-2009; Biological Evaluation of Medical Devices—Part 5: Tests for In Vitro Cytotoxicity. International Organization for Standardization: Geneva, Switzerland, 2009. Available online: https://nhiso.com/wp-content/uploads/2018/05/ISO-10993-5-2009.pdf (accessed on 25 March 2022).
- Stockert, J.C.; Horobin, R.W.; Colombo, L.L.; Blázquez-Castro, A. Tetrazolium Salts and Formazan Products in Cell Biology: Viability Assessment, Fluorescence Imaging, and Labeling Perspectives. Acta Histochem. 2018, 120, 159–167. [Google Scholar] [CrossRef]
- Da Violante, G.; Zerrouk, N.; Richard, I.; Provot, G.; Chaumeil, J.C.; Arnaud, P. Evaluation of the Cytotoxicity Effect of Dimethyl Sulfoxide (DMSO) on Caco2/TC7 Colon Tumor Cell Cultures. Biol. Pharm. Bull. 2002, 25, 1600–1603. [Google Scholar] [CrossRef]
- de Abreu Costa, L.; Henrique Fernandes Ottoni, M.; dos Santos, M.; Meireles, A.; Gomes de Almeida, V.; de Fátima Pereira, W.; Alves de Avelar-Freitas, B.; Eustáquio Alvim Brito-Melo, G. Dimethyl Sulfoxide (DMSO) Decreases Cell Proliferation and TNF-α, IFN-γ, and IL-2 Cytokines Production in Cultures of Peripheral Blood Lymphocytes. Molecules 2017, 22, 1789. [Google Scholar] [CrossRef]
- Singh, N.P.; McCoy, M.T.; Tice, R.R.; Schneider, E.L. A Simple Technique for Quantitation of Low Levels of DNA Damage in Individual Cells. Exp. Cell Res. 1988, 175, 184–191. [Google Scholar] [CrossRef]
- Singh, N.P.; Stephens, R.E. Microgel Electrophoresis: Sensitivity, Mechanisms, and DNA Electrostretching. Mutat. Res./DNA Repair 1997, 383, 167–175. [Google Scholar] [CrossRef]
- Lu, Y.; Liu, Y.; Yang, C. Evaluating In Vitro DNA Damage Using Comet Assay. J. Vis. Exp. 2017, 128, e56450. [Google Scholar] [CrossRef]
- Hartmann, A. Recommendations for Conducting the in Vivo Alkaline Comet Assay. Mutagenesis 2003, 18, 45–51. [Google Scholar] [CrossRef]
- Liao, W.; McNutt, M.A.; Zhu, W.-G. The Comet Assay: A Sensitive Method for Detecting DNA Damage in Individual Cells. Methods 2009, 48, 46–53. [Google Scholar] [CrossRef] [PubMed]
- Gyori, B.M.; Venkatachalam, G.; Thiagarajan, P.S.; Hsu, D.; Clement, M.-V. OpenComet: An Automated Tool for Comet Assay Image Analysis. Redox Biol. 2014, 2, 457–465. [Google Scholar] [CrossRef] [PubMed]
- Mosieniak, G.; Sliwinska, M.A.; Przybylska, D.; Grabowska, W.; Sunderland, P.; Bielak-Zmijewska, A.; Sikora, E. Curcumin-Treated Cancer Cells Show Mitotic Disturbances Leading to Growth Arrest and Induction of Senescence Phenotype. Int. J. Biochem. Cell Biol. 2016, 74, 33–43. [Google Scholar] [CrossRef]
- Lassmann, M.; Hänscheid, H.; Gassen, D.; Biko, J.; Meineke, V.; Reiners, C.; Scherthan, H. In Vivo Formation of γ-H2AX and 53BP1 DNA Repair Foci in Blood Cells After Radioiodine Therapy of Differentiated Thyroid Cancer. J. Nucl. Med. 2010, 51, 1318–1325. [Google Scholar] [CrossRef] [PubMed]
- Plappert-Helbig, U.; Libertini, S.; Frieauff, W.; Theil, D.; Martus, H.-J. Gamma-H2AX Immunofluorescence for the Detection of Tissue-Specific Genotoxicity in vivo: Gamma-H2AX Tissue-Specific Genotoxicity. Environ. Mol. Mutagen. 2019, 60, 4–16. [Google Scholar] [CrossRef] [PubMed]








| Cancer Cell Line | Compound | Alkaline Version | Neutral Version |
|---|---|---|---|
| HCT 116 | Control | 1.3 (0.5–2.1) | 2.9 (1.9–4) |
| MM129 IC50 | 10.7 * (4.7–28.1) | 19.4 * (13.3–29.2) | |
| MM129 2 × IC50 | 24.9 * (12.4–46.3) | 26.9 * (19.9–34.3) | |
| MM130 IC50 | 22.7 * (10.3–44) | 21.2 * (14.9–31.7) | |
| MM130 2 × IC50 | 25.6 * (14.1–41.3) | 30.6 * (24.8–36.1) | |
| MM131 IC50 | 31.5 * (19.8–51.3) | 15.8 * (11.7–23.1) | |
| MM131 2 × IC50 | 38.8 * (25.3–57) | 25.6 * (19.7–33.1) | |
| PC-3 | Control | 3.4 (2.2–4.8) | 1.3 * (0.4–2.3) |
| MM129 IC50 | 13.8 * (4–30.6) | 2.4 * (1.1–3.8) | |
| MM129 2 × IC50 | 21.8 * (10.4–38.2) | 7.3 * (5.1–10.3) | |
| MM130 IC50 | 12.4 * (4.2–33.4) | 1.5 * (0.4–3.1) | |
| MM130 2 × IC50 | 19.8 * (9.6–34) | 2.2 * (1.2–3.7) | |
| MM131 IC50 | 29.4 * (17.9–41) | 2.3 * (1.3–3.6) | |
| MM131 2 × IC50 | 40.3 * (28–53.2) | 4.3 * (2.8–6.3) | |
| BxPC-3 | Control | 1.6 * (0.5–2.6) | 2.5 * (0.4–4.4) |
| MM129 IC50 | 6.7 * (2.3–20.7) | 8.7 * (6.1–13.9) | |
| MM129 2 × IC50 | 20.7 * (10.5–27.4) | 10 * (6.9–13.9) | |
| MM130 IC50 | 9.1 * (2.2–25) | 7 * (4.7–12.2) | |
| MM130 2 × IC50 | 15.8 * (4.5–29.9) | 8.5 * (6.1–12.9) | |
| MM131 IC50 | 7.5 * (1.7–23.9) | 9.6 * (6.8–13.4) | |
| MM131 2 × IC50 | 17.4 * (2.5–27.7) | 9.7 * (7.4–12.9) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bukowski, K.; Marciniak, B.; Kciuk, M.; Mojzych, M.; Kontek, R. Pyrazolo[4,3-e]tetrazolo[1,5-b][1,2,4]triazine Sulfonamides as Novel Potential Anticancer Agents: Cytotoxic and Genotoxic Activities In Vitro. Molecules 2022, 27, 3761. https://doi.org/10.3390/molecules27123761
Bukowski K, Marciniak B, Kciuk M, Mojzych M, Kontek R. Pyrazolo[4,3-e]tetrazolo[1,5-b][1,2,4]triazine Sulfonamides as Novel Potential Anticancer Agents: Cytotoxic and Genotoxic Activities In Vitro. Molecules. 2022; 27(12):3761. https://doi.org/10.3390/molecules27123761
Chicago/Turabian StyleBukowski, Karol, Beata Marciniak, Mateusz Kciuk, Mariusz Mojzych, and Renata Kontek. 2022. "Pyrazolo[4,3-e]tetrazolo[1,5-b][1,2,4]triazine Sulfonamides as Novel Potential Anticancer Agents: Cytotoxic and Genotoxic Activities In Vitro" Molecules 27, no. 12: 3761. https://doi.org/10.3390/molecules27123761
APA StyleBukowski, K., Marciniak, B., Kciuk, M., Mojzych, M., & Kontek, R. (2022). Pyrazolo[4,3-e]tetrazolo[1,5-b][1,2,4]triazine Sulfonamides as Novel Potential Anticancer Agents: Cytotoxic and Genotoxic Activities In Vitro. Molecules, 27(12), 3761. https://doi.org/10.3390/molecules27123761