(±)-Cryptamides A–D, Four Pairs of Novel Dopamine Enantiomer Trimers from the Periostracum Cicadae
Abstract
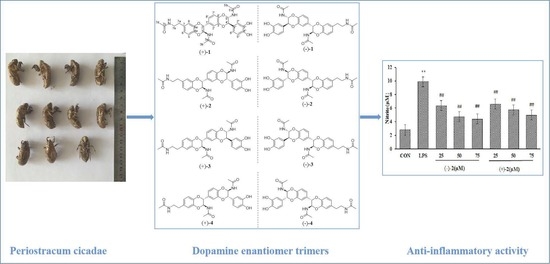
:1. Introduction
2. Results
2.1. Structure Elucidation
2.2. Nitric Oxide Inhibitory Activity
3. Materials and Methods
3.1. General Experimental Procedures
3.2. Plant Material
3.3. Extraction and Isolation
3.4. Compound Characterization
3.5. Computational Analysis
3.6. Cell Culture and Nitric Oxide Inhibitory Assay
3.7. Statistical Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Sample Availability
References
- China Pharmacopoeia Committee. Pharmacopoeia of People’s Republic of China; China Medico-Pharmaceutical Science & Technology Publishing House: Beijing, China, 2020; Volume 1, p. 385. [Google Scholar]
- Liu, H.; Yan, Y.M.; Wang, S.X.; Zhang, Y.; Cheng, Y.X. Cicadamides A and B, N-Acetyldopamine Dimers from the Insect Periostracum cicadae. Nat. Prod. Com. 2019, 14, 1–6. [Google Scholar] [CrossRef] [Green Version]
- Hsieh, M.T.; Peng, W.H.; Yeh, F.T.; Tsai, H.Y. Studies on the anticonvulsive, sedative and hypothermic effects of Periostracum Cicadae extracts. J. Ethnopharmacol. 1991, 35, 83–90. [Google Scholar] [CrossRef]
- Yang, L.; Wang, Y.; Nuerbiye, A.; Cheng, P.; Wang, J.H.; Kasimu, R.; Li, H. Effects of Periostracum Cicadae on Cytokines and Apoptosis Regulatory Proteins in an IgA Nephropathy Rat Model. Int. J. Mol. Sci. 2018, 19, 1599. [Google Scholar] [CrossRef] [Green Version]
- Xu, M.Z.; Lee, W.S.; Han, J.M.; Oh, H.W.; Park, D.S.; Tian, G.R.; Jeong, T.S.; Park, H.Y. Antioxidant and anti-inflammatory activities of N-acetyldopamine dimers from Periostracum Cicadae. Med. Chem. 2006, 14, 7826–7834. [Google Scholar] [CrossRef]
- Thapa, P.; Gu, Y.; Kil, Y.S.; Baek, S.C.; Kim, H.K.; Han, A.R.; Seo, E.K.; Choi, H.; Chang, J.H.; Nam, J.W. N-Acetyldopamine derivatives from Periostracum Cicadae and their regulatory activities on Th1 and Th17 cell differentiation. Bioorg. Chem. 2020, 102, 104095. [Google Scholar] [CrossRef]
- Lu, Y.; Guo, Y.L.; Wang, H.Y.; Zhang, K.; Zhu, Y.; Zhao, W.; Wang, H.; Wang, J.H. N-acetyldopamine derivatives from Periostracum Cicadae. Phytochem. Lett. 2015, 11, 275–279. [Google Scholar] [CrossRef]
- Noda, N.; Kubota, S.; Miyata, Y.; Miyahara, K. Optically active N-acetyldopamine dimer of the crude drug “Zentai”, the cast-off shell of the Cicada, Cryptotympana sp. Chem. Pharm. Bull. 2000, 48, 1749–1752. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, L.; Li, G.Y.; Wang, H.Y.; Zhang, K.; Zhu, Y.; Zhao, W.; Wang, H.; Wang, J.H. Five new N-acetyldopamine dimers from Periostracum Cicadae. Phytochem. Lett. 2016, 16, 97–102. [Google Scholar] [CrossRef]
- Lee, W.; Lee, H.; Kim, M.A.; Choi, J.; Kim, K.M.; Hwang, J.; Na, M.; Bae, J.S. Evaluation of novel factor Xa inhibitors from Oxya chinensis sinuosa with anti-platelet aggregation activity. Sci. Rep. 2017, 7, 7934. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, J.; Li, Y.P.; Qin, F.Y.; Yan, Y.M.; Zhang, H.X.; Cheng, Y.X. Racemic xanthine and dihydroxydopamine conjugates from Cyclopelta parva and their COX- 2 inhibitory activity. Fitoterapia 2020, 142, 104534. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Li, G.Y.; Li, Q.R.; Wang, J.H.J. Two new N-acetyldopamine tetrapolymers from Periostracum Cicadae. Asian Nat. Prod. Res. 2012, 14, 204–209. [Google Scholar] [CrossRef] [PubMed]
- Zhu, H.J.; Yan, Y.M.; Tu, Z.C.; Luo, J.F.; Liang, R.; Yang, T.H.; Cheng, Y.X.; Wang, S.M. Compounds from Polyphaga plancyi and their inhibitory activities against JAK3 and DDR1 kinases. Fitoterapia 2016, 114, 163–167. [Google Scholar] [CrossRef] [PubMed]
- Lu, J.; Sun, Q.; Tu, Z.C.; Lv, Q.; Shui, P.X.; Cheng, Y.X. Identification of N-Acetyldopamine Dimers from the Dung Beetle Catharsius molossus and Their COX-1 and COX-2 Inhibitory Activities. Molecules 2015, 20, 15589–15596. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shi, Y.N.; Tu, Z.C.; Wang, X.L.; Yan, Y.M.; Fang, P.; Zuo, Z.L.; Hou, B.; Yang, T.H.; Cheng, Y.X. Bioactive compounds from the insect Aspongopus chinensis. Bioorg. Med. Chem. Lett. 2014, 24, 5164–5169. [Google Scholar] [CrossRef] [PubMed]





| No. | 1 | 2 | 3 | 4 | ||||
|---|---|---|---|---|---|---|---|---|
| δCa/ppm | δH/ppm (b J in Hz) | δC/ppm | δH/ppm (J in Hz) | δC/ppm | δH/ppm (J in Hz) | δC/ppm | δH/ppm (J in Hz) | |
| 2 | 78.3 | 4.75 (t, 7.0) | 78.2 | 4.77 (d, 7.0) | 78.3 | 4.77 (d, 7.0) | 78.3 | 4.76 (d, 7.0) |
| 3 | 78.4 | 5.71 (t, 7.0) | 78.3 | 5.74 (d, 7.0) | 78.4 | 5.72 (d, 7.0) | 78.3 | 5.74 (d, 7.0) |
| 4a | 142.8 | 142.1 | 142.1 | 142.8 | ||||
| 5 | 118.0 | 6.88 (d, 8.5) | 118.1 | 6.87 (overlap) | 118.0 | 6.77 (overlap) | 118.0 | 6.77 (overlap) |
| 6 | 123.2 | 6.75 (overlap) | 123.4 | 6.75 (overlap) | 134.3 | 134.5 | ||
| 7 | 134.5 | 134.3 | 123.4 | 6.75 (overlap) | 123.2 | 6.75 (overlap) | ||
| 8 | 118.2 | 6.77 (overlap) | 118.1 | 6.77 (overlap) | 118.1 | 6.83 (overlap) | 118.2 | 6.84 (overlap) |
| 8a | 144.5 | 144.4 | 144.3 | 144.4 | ||||
| 1′ | 131.0 | 131.0 | 130.1 | 131.0 | ||||
| 2′ | 117.9 | 6.95 (d, 1.5) | 118.0 | 6.95 (d, 1.5) | 118.0 | 6.95 (d, 1.5) | 117.9 | 6.95 (d, 1.5) |
| 3′ | 144.3 | 144.2 | 144.1 | 144.2 | ||||
| 4′ | 77.9 | 4.75 (d, 7.0) | 77.9 | 4.73 (d, 7.0) | 78.0 | 4.74 (d, 7.0) | 77.8 | 4.73 (d, 7.0) |
| 5′ | 78.3 | 5.71 (d, 7.0) | 78.2 | 5.68 (d, 7.0) | 78.2 | 5.70 (d, 7.0) | 78.3 | 5.69 (d, 7.0) |
| 6′ | 143.5 | 144.1 | 144.4 | 143.5 | ||||
| 7′ | 117.3 | 6.86 (overlap) | 117.4 | 6.82 (overlap) | 117.3 | 6.82 (overlap) | 117.3 | 6.84 (overlap) |
| 8′ | 122.4 | 6.96 (overlap) | 122.2 | 6.96 (overlap) | 122.4 | 6.96 (overlap) | 122.2 | 6.96 (overlap) |
| 1″ | 128.6 | 128.5 | 128.5 | 128.6 | ||||
| 2″ | 115.5 | 6.83 (d, 1.5) | 115.6 | 6.80 (s) | 115.5 | 6.81 (s) | 115.5 | 6.84 (d, 1.5) |
| 3″ | 146.5 | 146.5 | 146.5 | 146.5 | ||||
| 4″ | 147.2 | 147.2 | 147.2 | 147.3 | ||||
| 5″ | 116.2 | 7.02 (overlap) | 116.1 | 7.01 (overlap) | 116.2 | 7.02 (overlap) | 116.1 | 7.01 (overlap) |
| 6″ | 120.6 | 6.73 (overlap) | 120.6 | 6.73 (overlap) | 120.6 | 6.73 (overlap) | 120.6 | 6.73 (overlap) |
| 3a | 173.3 | 173.3 | 173.3 | 173.3 | ||||
| 3b | 22.6 | 1.90 (s) | 22.6 | 1.90 (s) | 22.6 | 1.90 (s) | 22.6 | 1.90 (s) |
| 5′a | 173.3 | 173.3 | 173.3 | 173.3 | ||||
| 5′b | 22.6 | 1.89 (s) | 22.6 | 1.89 (s) | 22.6 | 1.89 (s) | 22.6 | 1.89 (s) |
| 7a/6a | 35.8 | 2.70 (t, 7.0) | 35.8 | 2.70 (t, 7.0) | 35.8 | 2.70 (t, 7.0) | 35.8 | 2.70 (t, 7.0) |
| 7b/6b | 42.1 | 3.35 (t, 7.5) | 42.1 | 3.35 (t, 7.5) | 42.1 | 3.35 (t, 7.5) | 42.1 | 3.35 (t, 7.5) |
| 7c/6c | 173.3 | 173.2 | 173.2 | 173.2 | ||||
| 7d/6d | 22.5 | 1.88 (s) | 22.5 | 1.88 (s) | 22.5 | 1.88 (s) | 22.5 | 1.88 (s) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Luo, J.; Wei, W.; Wang, P.; Guo, T.; Chen, S.; Zhang, L.; Feng, S. (±)-Cryptamides A–D, Four Pairs of Novel Dopamine Enantiomer Trimers from the Periostracum Cicadae. Molecules 2022, 27, 6707. https://doi.org/10.3390/molecules27196707
Luo J, Wei W, Wang P, Guo T, Chen S, Zhang L, Feng S. (±)-Cryptamides A–D, Four Pairs of Novel Dopamine Enantiomer Trimers from the Periostracum Cicadae. Molecules. 2022; 27(19):6707. https://doi.org/10.3390/molecules27196707
Chicago/Turabian StyleLuo, Junjian, Wenjun Wei, Pan Wang, Tao Guo, Suiqing Chen, Liping Zhang, and Shuying Feng. 2022. "(±)-Cryptamides A–D, Four Pairs of Novel Dopamine Enantiomer Trimers from the Periostracum Cicadae" Molecules 27, no. 19: 6707. https://doi.org/10.3390/molecules27196707
APA StyleLuo, J., Wei, W., Wang, P., Guo, T., Chen, S., Zhang, L., & Feng, S. (2022). (±)-Cryptamides A–D, Four Pairs of Novel Dopamine Enantiomer Trimers from the Periostracum Cicadae. Molecules, 27(19), 6707. https://doi.org/10.3390/molecules27196707