Presence of β-Turn Structure in Recombinant Spider Silk Dissolved in Formic Acid Revealed with NMR
Abstract
:1. Introduction
2. Results and Discussion
2.1. 1H, 13C, and 15N Assignments of the Repetitive Domain in RSP in Formic Acid
2.2. Secondary Structure of the Repetitive Sequence in RSP in Formic Acid
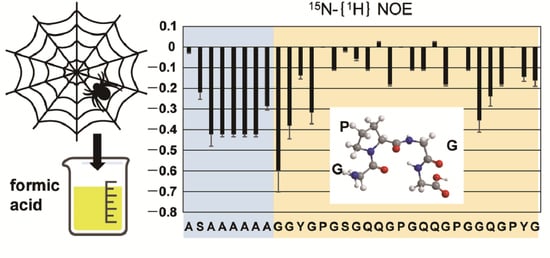
2.3. Dynamics of the Repetitive Sequence in RSP in Formic Acid by 15N-{1H} Steady-State NOE Measurement
2.4. Solvent Effect of Formic Acid on RSP Structure
2.5. Formylation of RSP Occurred in Formic Acid
2.6. Secondary Structure of the RSP Film in the Solid State Prepared from Formic Acid
3. Materials and Methods
3.1. Preparation of Recombinant Spider Silk Protein, RSP
3.2. Solution NMR Measurements
3.3. Solid-State NMR Measurements
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
References
- Huang, W.; Ling, S.; Li, C.; Omenetto, F.G.; Kaplan, D.L. Silkworm silk-based materials and devices generated using bio-nanotechnology. Chem. Soc. Rev. 2018, 47, 6486–6504. [Google Scholar] [CrossRef]
- Koh, L.-D.; Cheng, Y.; Teng, C.-P.; Khin, Y.-W.; Loh, X.-J.; Tee, S.-Y.; Low, M.; Ye, E.; Yu, H.-D.; Zhang, Y.-W.; et al. Structures, mechanical properties and applications of silk fibroin materials. Prog. Polym. Sci. 2015, 46, 86–110. [Google Scholar] [CrossRef]
- Kundu, B.; Rajkhowa, R.; Kundu, S.C.; Wang, X. Silk fibroin biomaterials for tissue regenerations. Adv. Drug Deliv. Rev. 2013, 65, 457–470. [Google Scholar] [CrossRef]
- Asakura, T.; Suzuki, Y.; Kametani, S. Silk. In Encyclopedia of Polymer Science and Technology; Wiley: Hoboken, NJ, USA, 2018. [Google Scholar] [CrossRef]
- Koeppel, A.; Holland, C. Progress and Trends in Artificial Silk Spinning: A Systematic Review. ACS Biomater. Sci. Eng. 2017, 3, 226–237. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Trabbic, K.A.; Yager, P. Comparative Structural Characterization of Naturally- and Synthetically-Spun Fibers of Bombyx mori Fibroin. Macromolecules 1998, 31, 462–471. [Google Scholar] [CrossRef]
- Liivak, O.; Blye, A.; Shah, N.; Jelinski, L.W. A Microfabricated Wet-Spinning Apparatus To Spin Fibers of Silk Proteins. StructureProperty Correlations. Macromolecules 1998, 31, 2947–2951. [Google Scholar] [CrossRef]
- Seidel, A.; Liivak, O.; Jelinski, L.W. Artificial Spinning of Spider Silk. Macromolecules 1998, 31, 6733–6736. [Google Scholar] [CrossRef]
- Zhao, C.; Yao, J.; Masuda, H.; Raghuvansh, K.; Asakura, T. Structural characterization and artificial fiber formation of Bombyx mori silk fibroin in hexafluoro-iso-propanol solvent system. Biopolymers 2003, 69, 253–259. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Z.; Kikuchi, Y.; Kojima, K.; Tamura, T.; Kuwabara, N.; Nakamura, T.; Asakura, T. Mechanical Properties of Regenerated Bombyx mori Silk Fibers and Recombinant Silk Fibers Produced by Transgenic Silkworms. J. Biomater. Sci. Polym. Ed. 2010, 21, 395–411. [Google Scholar] [CrossRef]
- Suzuki, Y.; Gerig, J.T.; Asakura, T. NMR Study of Interactions between Silk Model Peptide and Fluorinated Alcohols for Preparation of Regenerated Silk Fiber. Macromolecules 2010, 43, 2364–2370. [Google Scholar] [CrossRef]
- Yao, J.; Masuda, H.; Zhao, C.; Asakura, T. Artificial spinning and characterization of silk fiber from Bombyx mori silk fibroin in hexafluoroacetone hydrate. Macromolecules 2002, 35, 6–9. [Google Scholar] [CrossRef]
- Ha, S.-W.; Tonelli, A.E.; Hudson, S.M. Structural Studies of Bombyx mori Silk Fibroin during Regeneration from Solutions and Wet Fiber Spinning. Biomacromolecules 2005, 6, 1722–1731. [Google Scholar] [CrossRef]
- Marsano, E.; Corsini, P.; Arosio, C.; Boschi, A.; Mormino, M.; Freddi, G. Wet spinning of Bombyx mori silk fibroin dissolved in N-methyl morpholine N-oxide and properties of regenerated fibres. Int. J. Biol. Macromol. 2005, 37, 179–188. [Google Scholar] [CrossRef] [PubMed]
- Hijirida, D.H.; Do, K.G.; Michal, C.; Wong, S.; Zax, D.; Jelinski, L.W. 13C NMR of Nephila clavipes major ampullate silk gland. Biophys. J. 1996, 71, 3442–3447. [Google Scholar] [CrossRef] [Green Version]
- Um, I.C.; Kweon, H.; Park, Y.H.; Hudson, S. Structural characteristics and properties of the regenerated silk fibroin prepared from formic acid. Int. J. Biol. Macromol. 2001, 29, 91–97. [Google Scholar] [CrossRef]
- Um, I.C.; Kweon, H.Y.; Lee, K.G.; Park, Y.H. The role of formic acid in solution stability and crystallization of silk protein polymer. Int. J. Biol. Macromol. 2003, 33, 203–213. [Google Scholar] [CrossRef] [PubMed]
- Yao, J.; Ohgo, K.; Sugino, R.; Kishore, R.; Asakura, T. Structural Analysis of Bombyx mori Silk Fibroin Peptides with Formic Acid Treatment Using High-Resolution Solid-State 13C NMR Spectroscopy. Biomacromolecules 2004, 5, 1763–1769. [Google Scholar] [CrossRef] [PubMed]
- Ghanaati, S.; Orth, C.; Unger, R.E.; Barbeck, M.; Webber, M.J.; Motta, A.; Migliaresi, C.; James Kirkpatrick, C. Fine-tuning scaffolds for tissue regeneration: Effects of formic acid processing on tissue reaction to silk fibroin. J. Tissue Eng. Regen. Med. 2010, 4, 464–472. [Google Scholar] [CrossRef]
- Zhang, F.; Lu, Q.; Yue, X.; Zuo, B.; Qin, M.; Li, F.; Kaplan, D.L.; Zhang, X. Regeneration of high-quality silk fibroin fiber by wet spinning from CaCl2–formic acid solvent. Acta Biomater. 2015, 12, 139–145. [Google Scholar] [CrossRef]
- Zhang, F.; You, X.; Dou, H.; Liu, Z.; Zuo, B.; Zhang, X. Facile Fabrication of Robust Silk Nanofibril Films via Direct Dissolution of Silk in CaCl2–Formic Acid Solution. ACS Appl. Mater. Interfaces 2015, 7, 3352–3361. [Google Scholar] [CrossRef]
- Liu, Q.; Wang, F.; Gu, Z.; Ma, Q.; Hu, X. Exploring the Structural Transformation Mechanism of Chinese and Thailand Silk Fibroin Fibers and Formic-Acid Fabricated Silk Films. Int. J. Mol. Sci. 2018, 19, 3309. [Google Scholar] [CrossRef] [Green Version]
- Xue, Y.; Wang, F.; Torculas, M.; Lofland, S.; Hu, X. Formic Acid Regenerated Mori, Tussah, Eri, Thai, and Muga Silk Materials: Mechanism of Self-Assembly. ACS Biomater. Sci. Eng. 2019, 5, 6361–6373. [Google Scholar] [CrossRef] [PubMed]
- Gosline, J.M.; DeMont, M.E.; Denny, M.W. The structure and properties of spider silk. Endeavour 1986, 10, 37–43. [Google Scholar] [CrossRef]
- Xu, M.; Lewis, R. V Structure of a protein superfiber: Spider dragline silk. Proc. Natl. Acad. Sci. USA 1990, 87, 7120–7124. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Termonia, Y. Molecular Modeling of Spider Silk Elasticity. Macromolecules 1994, 27, 7378–7381. [Google Scholar] [CrossRef]
- Cunniff, P.M.; Fossey, S.A.; Auerbach, M.A.; Song, J.W.; Kaplan, D.L.; Adams, W.W.; Eby, R.K.; Mahoney, D.; Vezie, D.L. Mechanical and thermal properties of dragline silk from the spider Nephila clavipes. Polym. Adv. Technol. 1994, 5, 401–410. [Google Scholar] [CrossRef]
- Gosline, J.M.; Guerette, P.A.; Ortlepp, C.S.; Savage, K.N. The mechanical design of spider silks: From fibroin sequence to mechanical function. J. Exp. Biol. 1999, 202, 3295–3303. [Google Scholar] [CrossRef]
- Vollrath, F.; Knight, D.P. Liquid crystalline spinning of spider silk. Nature 2001, 410, 541–548. [Google Scholar] [CrossRef]
- Eisoldt, L.; Smith, A.; Scheibel, T. Decoding the secrets of spider silk. Mater. Today 2011, 14, 80–86. [Google Scholar] [CrossRef]
- Blackledge, T.A.; Pérez-Rigueiro, J.; Plaza, G.R.; Perea, B.; Navarro, A.; Guinea, G.V.; Elices, M. Sequential origin in the high performance properties of orb spider dragline silk. Sci. Rep. 2012, 2, 782–787. [Google Scholar] [CrossRef] [Green Version]
- Asakura, T.; Miller, T. Biotechnology of Silk; Asakura, T., Miller, T., Eds.; Biologically-Inspired Systems; Springer: Dordrecht, The Netherlands, 2014; Volume 5, ISBN 978-94-007-7118-5. [Google Scholar]
- Tokareva, O.; Jacobsen, M.; Buehler, M.; Wong, J.; Kaplan, D.L. Structure–function–property–design interplay in biopolymers: Spider silk. Acta Biomater. 2014, 10, 1612–1626. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Asakura, T.; Tasei, Y.; Aoki, A.; Nishimura, A. Mixture of Rectangular and Staggered Packing Arrangements of Polyalanine Region in Spider Dragline Silk in Dry and Hydrated States as Revealed by 13C NMR and X-ray Diffraction. Macromolecules 2018, 51, 1058–1068. [Google Scholar] [CrossRef]
- Asakura, T.; Nishimura, A.; Tasei, Y. Determination of Local Structure of 13C Selectively Labeled 47-mer Peptides as a Model for Gly-Rich Region of Nephila clavipes Dragline Silk Using a Combination of 13C Solid-State NMR and MD Simulation. Macromolecules 2018, 51, 3608–3619. [Google Scholar] [CrossRef]
- Oktaviani, N.A.; Matsugami, A.; Malay, A.D.; Hayashi, F.; Kaplan, D.L.; Numata, K. Conformation and dynamics of soluble repetitive domain elucidates the initial β-sheet formation of spider silk. Nat. Commun. 2018, 9, 2121. [Google Scholar] [CrossRef] [PubMed]
- Yarger, J.L.; Cherry, B.R.; van der Vaart, A. Uncovering the structure–function relationship in spider silk. Nat. Rev. Mater. 2018, 3, 18008. [Google Scholar] [CrossRef]
- Asakura, T.; Nishimura, A.; Aoki, A.; Naito, A. Packing Structure of Antiparallel β-Sheet Polyalanine Region in a Sequential Model Peptide of Nephila clavipes Dragline Silk Studied Using 13C Solid-State NMR and MD Simulation. Biomacromolecules 2019, 20, 3884–3894. [Google Scholar] [CrossRef] [PubMed]
- Yazawa, K.; Malay, A.D.; Masunaga, H.; Norma-Rashid, Y.; Numata, K. Simultaneous effect of strain rate and humidity on the structure and mechanical behavior of spider silk. Commun. Mater. 2020, 1, 10. [Google Scholar] [CrossRef] [Green Version]
- Malay, A.D.; Suzuki, T.; Katashima, T.; Kono, N.; Arakawa, K.; Numata, K. Spider silk self-assembly via modular liquid-liquid phase separation and nanofibrillation. Sci. Adv. 2020, 6, eabb6030. [Google Scholar] [CrossRef]
- Asakura, T. Structure and Dynamics of Spider Silk Studied with Solid-State Nuclear Magnetic Resonance and Molecular Dynamics Simulation. Molecules 2020, 25, 2634. [Google Scholar] [CrossRef]
- Work, R.W.; Morosoff, N. A Physico-Chemical Study of the Supercontraction of Spider Major Ampullate Silk Fibers. Text. Res. J. 1982, 52, 349–356. [Google Scholar] [CrossRef]
- Work, R.W. Viscoelastic Behaviour and Wet Supercontraction of Major Ampullate Silk Fibres of Certain Orb-Web-Building Spiders (Araneae). J. Exp. Biol. 1985, 118, 379–404. [Google Scholar] [CrossRef]
- Fu, C.; Shao, Z.; Fritz, V. Animal silks: Their structures, properties and artificial production. Chem. Commun. 2009, 37, 6515–6529. [Google Scholar] [CrossRef]
- Savage, K.N.; Gosline, J.M. The effect of proline on the network structure of major ampullate silks as inferred from their mechanical and optical properties. J. Exp. Biol. 2008, 211, 1937–1947. [Google Scholar] [CrossRef] [Green Version]
- Liu, Y.; Sponner, A.; Porter, D.; Vollrath, F. Proline and Processing of Spider Silks. Biomacromolecules 2008, 9, 116–121. [Google Scholar] [CrossRef]
- Tasei, Y.; Nishimura, A.; Suzuki, Y.Y.; Sato, T.K.; Sugahara, J.; Asakura, T. NMR Investigation about Heterogeneous Structure and Dynamics of Recombinant Spider Silk in the Dry and Hydrated States. Macromolecules 2017, 50, 8117–8128. [Google Scholar] [CrossRef]
- Guerette, P.A.; Ginzinger, D.G.; Weber, B.H.; Gosline, J.M. Silk properties determined by gland-specific expression of a spider fibroin gene family. Science 1996, 272, 112–115. [Google Scholar] [CrossRef]
- Rising, A.; Widhe, M.; Johansson, J.; Hedhammar, M. Spider silk proteins: Recent advances in recombinant production, structure–function relationships and biomedical applications. Cell. Mol. Life Sci. 2011, 68, 169–184. [Google Scholar] [CrossRef]
- Tamara, A.B.; DeSimone, E.; Scheibel, T. Biomedical Applications of Recombinant Silk-Based Materials. Adv. Mater. 2018, 30, 1704636. [Google Scholar] [CrossRef]
- Salehi, S.; Koeck, K.; Scheibel, T. Spider Silk for Tissue Engineering Applications. Molecules 2020, 25, 737. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Klunk, W.E.; Pettegrew, J.W. Alzheimer’s β-Amyloid Protein Is Covalently Modified when Dissolved in Formic Acid. J. Neurochem. 1990, 54, 2050–2056. [Google Scholar] [CrossRef] [PubMed]
- Klunk, W.E.; Xu, C.-J.; Pettegrew, J.W. NMR Identification of the Formic Acid-Modified Residue in Alzheimer’s Amyloid Protein. J. Neurochem. 1994, 62, 349–354. [Google Scholar] [CrossRef] [PubMed]
- Zheng, S.; Doucette, A.A. Preventing N- and O-formylation of proteins when incubated in concentrated formic acid. Proteomics 2016, 16, 1059–1068. [Google Scholar] [CrossRef] [PubMed]
- Lenčo, J.; Khalikova, M.A.; Švec, F. Dissolving Peptides in 0.1% Formic Acid Brings Risk of Artificial Formylation. J. Proteome Res. 2020, 19, 993–999. [Google Scholar] [CrossRef]
- Shen, Y.; Bax, A. Protein backbone and sidechain torsion angles predicted from NMR chemical shifts using artificial neural networks. J. Biomol. NMR 2013, 56, 227–241. [Google Scholar] [CrossRef] [Green Version]
- Suzuki, Y.Y.; Yamazaki, T.; Aoki, A.; Shindo, H.; Asakura, T. NMR Study of the Structures of Repeated Sequences, GAGXGA (X = S, Y, V), in Bombyx mori Liquid Silk. Biomacromolecules 2014, 15, 104–112. [Google Scholar] [CrossRef] [PubMed]
- Wishart, D.S. Interpreting protein chemical shift data. Prog. Nucl. Magn. Reson. Spectrosc. 2011, 58, 62–87. [Google Scholar] [CrossRef]
- Jenkins, J.E.; Holland, G.P.; Yarger, J.L. High resolution magic angle spinning NMR investigation of silk protein structure within major ampullate glands of orb weaving spiders. Soft Matter 2012, 8, 1947–1954. [Google Scholar] [CrossRef]
- Xu, D.; Yarger, J.L.; Holland, G.P. Exploring the backbone dynamics of native spider silk proteins in Black Widow silk glands with solution-state NMR spectroscopy. Polymer 2014, 55, 3879–3885. [Google Scholar] [CrossRef]
- Jenkins, J.E.; Creager, M.S.; Butler, E.B.; Lewis, R.V.; Yarger, J.L.; Holland, G.P. Solid-state NMR evidence for elastin-like β-turn structure in spider dragline silk. Chem. Commun. 2010, 46, 6714–6716. [Google Scholar] [CrossRef]
- Aluigi, A.; Zoccola, M.; Vineis, C.; Tonin, C.; Ferrero, F.; Canetti, M. Study on the structure and properties of wool keratin regenerated from formic acid. Int. J. Biol. Macromol. 2007, 41, 266–273. [Google Scholar] [CrossRef]
- Ha, S.-W.; Asakura, T.; Kishore, R. Distinctive influence of two hexafluoro solvents on the structural stabilization of Bombyx mori silk fibroin protein and its derived peptides: 13C NMR and CD studies. Biomacromolecules 2006, 7, 18–23. [Google Scholar] [CrossRef] [PubMed]
- Delaglio, F.; Grzesiek, S.; Vuister, G.W.; Zhu, G.; Pfeifer, J.; Bax, A. NMRPipe: A multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR 1995, 6, 277–293. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, N.; Harano, Y.; Tochio, N.; Nakatani, E.; Kigawa, T.; Yokoyama, S.; Mading, S.; Ulrich, E.L.; Markley, J.L.; Akutsu, H.; et al. An automated system designed for large scale NMR data deposition and annotation: Application to over 600 assigned chemical shift data entries to the BioMagResBank from the Riken Structural Genomics/Proteomics Initiative internal database. J. Biomol. NMR 2012, 53, 311–320. [Google Scholar] [CrossRef] [PubMed] [Green Version]








| Residue Number | Amino Acid | N | NH | HA | CA | CO | CB |
|---|---|---|---|---|---|---|---|
| 1 | Ala | 123.0 | 7.87 | 4.61 | 51.9 | 176.9 | 18.2 |
| 2 | Ser | 112.6 | 8.13 | 4.91 | 54.5 | 172.2 | 64.1 |
| 3 | Ala | 124.4 | 7.99 | 4.51 | 52.0 | 176.6 | 17.9 |
| 4~7 | Ala | 122.0 | 7.88 | 4.51 | 51.8 | 177.1 | 18.2 |
| 8 | Ala | 122.2 | 7.89 | 4.61 | 51.8 | 177.1 | 18.2 |
| 9 | Gly | 106.9 | 7.97 | 4.18 | 44.5 | 173.9 | - |
| 10 | Gly | 107.6 | 7.91 | 4.18 | 44.4 | 173.0 | - |
| 11 | Tyr | 119.1 | 8.01 | 4.87 | 56.9 | 175.1 | 38.3 |
| 12 | Gly | 110.0 | 7.93 | - | 43.8 | 171.2 | - |
| 13 | Pro | - | - | 4.73 | 62.8 | 176.7 | 31.0 |
| 14 | Gly | 108.0 | 8.10 | 4.28 | 44.5 | 173.4 | - |
| 15 | Ser | 113.4 | 8.12 | 5.04 | 54.3 | 172.8 | 64.5 |
| 16 | Gly | 109.0 | 8.18 | 4.18 | 44.7 | 173.3 | - |
| 17 | Gln | 118.9 | 8.01 | 4.69 | 54.9 | 174.7 | 28.7 |
| 18 | Gln | 120.3 | 8.18 | 4.74 | 54.8 | 174.8 | 28.9 |
| 19 | Gly | 109.6 | 8.07 | - | 43.9 | 171.2 | - |
| 20 | Pro | - | - | 4.69 | 62.8 | 176.7 | 31.0 |
| 21 | Gly | 108.0 | 8.10 | 4.28 | 44.5 | 173.4 | - |
| 22 | Gln | 118.9 | 8.01 | 4.69 | 54.9 | 174.7 | 28.7 |
| 23 | Gln | 120.3 | 8.18 | 4.74 | 54.8 | 174.8 | 28.9 |
| 24 | Gly | 109.6 | 8.07 | - | 43.9 | 171.2 | - |
| 25 | Pro | - | - | 4.69 | 62.8 | 176.7 | 31.0 |
| 26 | Gly | 108.0 | 8.10 | 4.28 | 44.5 | 173.4 | - |
| 27 | Gly | 107.7 | 7.98 | 4.25 | 44.5 | 173.4 | - |
| 28 | Gln | 118.6 | 8.04 | 4.74 | 54.8 | 173.4 | 28.6 |
| 29 | Gly | 109.6 | 8.07 | - | 43.9 | 171.2 | - |
| 30 | Pro | - | - | 4.63 | 62.6 | 175.6 | 30.9 |
| 31 | Tyr | 119.2 | 7.93 | 4.92 | 56.6 | 175.0 | 38.1 |
| 32 | Gly | 110.0 | 7.91 | - | 43.8 | 171.2 | - |
| Φi | Ψi | Φi + 1 | Ψi + 1 | Φi + 2 | |
|---|---|---|---|---|---|
| Type I β-turn | −65 ± 12 | −24 ± 13 | −93 ± 16 | ||
| Type II β-turn | −61 ± 13 | 136 ± 11 | 80 ± 16 | ||
| G12P13G14 | 105 | −177 | −65 | 148 | −124 |
| G19P20G21 | −88 | −180 | −63 | 146 | 73 |
| G24P25G26 | −87 | −178 | −66 | 148 | 111 |
| G29P30Y31 | 132 | −175 | −70 | 151 | −85 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Suzuki, Y.; Higashi, T.; Yamamoto, T.; Okamura, H.; Sato, T.K.; Asakura, T. Presence of β-Turn Structure in Recombinant Spider Silk Dissolved in Formic Acid Revealed with NMR. Molecules 2022, 27, 511. https://doi.org/10.3390/molecules27020511
Suzuki Y, Higashi T, Yamamoto T, Okamura H, Sato TK, Asakura T. Presence of β-Turn Structure in Recombinant Spider Silk Dissolved in Formic Acid Revealed with NMR. Molecules. 2022; 27(2):511. https://doi.org/10.3390/molecules27020511
Chicago/Turabian StyleSuzuki, Yu, Takanori Higashi, Takahiro Yamamoto, Hideyasu Okamura, Takehiro K. Sato, and Tetsuo Asakura. 2022. "Presence of β-Turn Structure in Recombinant Spider Silk Dissolved in Formic Acid Revealed with NMR" Molecules 27, no. 2: 511. https://doi.org/10.3390/molecules27020511
APA StyleSuzuki, Y., Higashi, T., Yamamoto, T., Okamura, H., Sato, T. K., & Asakura, T. (2022). Presence of β-Turn Structure in Recombinant Spider Silk Dissolved in Formic Acid Revealed with NMR. Molecules, 27(2), 511. https://doi.org/10.3390/molecules27020511