Uncovering the Mechanisms of Active Components from Toad Venom against Hepatocellular Carcinoma Using Untargeted Metabolomics
Abstract
:1. Introduction
2. Results
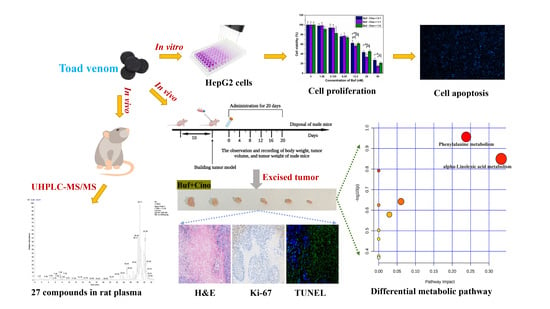
2.1. In Vivo Compound Identification of Toad Venom in Rat Plasma
2.2. In Vitro Antitumor Effect of Buf and Cino
2.3. In Vivo Antitumor Effect of Buf and Cino
2.4. Nontargeted Metabolomics Analysis of Tumor Tissues
2.4.1. Stability Test of Tumor Metabolomic Method
2.4.2. Identification of Differential Metabolites
2.4.3. Hierarchical Cluster Analysis of Differential Metabolites
2.4.4. Pathway Analysis of Differential Metabolites
3. Discussion
4. Materials and Methods
4.1. Materials and Reagents
4.2. Cells and Animals
4.3. In Vivo Compound Identification of Toad Venom in Rat Plasma
4.4. Cell Counting Kit-8 (CCK-8) Assay
4.5. Hoechst 33342 Staining Assay
4.6. In Vivo Antitumor Efficacy
4.7. Nontargeted Metabolomics Analysis of Tumor Tissues
4.7.1. Metabolites Extraction
4.7.2. UHPLC-Q-Exactive Orbitrap MS Analysis
4.7.3. MS Data Processing and Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Wei, L.; Wang, Z.; Jing, N.; Lu, Y.; Yang, J.; Xiao, H.; Guo, H.; Sun, S.; Li, M.; Zhao, D.; et al. Frontier progress of the combination of modern medicine and traditional Chinese medicine in the treatment of hepatocellular carcinoma. Chin. Med. 2022, 17, 90. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.; Chen, F.; Zhong, Z.; Tan, H.Y.; Wang, N.; Liu, Y.; Fang, X.; Yang, T.; Feng, Y. Interpreting the Pharmacological Mechanisms of Huachansu Capsules on Hepatocellular Carcinoma Through Combining Network Pharmacology and Experimental Evaluation. Front. Pharmacol. 2020, 11, 414. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Bao, H. Anti-tumor effect of Inonotus hispidus petroleum ether extract in H22 tumor-bearing mice and analysis its mechanism by untargeted metabonomic. J. Ethnopharmacol. 2022, 285, 114898. [Google Scholar] [CrossRef] [PubMed]
- Fan, Y.; Ma, Z.; Zhao, L.; Wang, W.; Gao, M.; Jia, X.; Ouyang, H.; He, J. Anti-tumor activities and mechanisms of Traditional Chinese medicines formulas: A review. Biomed. Pharmacother. 2020, 132, 110820. [Google Scholar] [CrossRef] [PubMed]
- Wei, W.L.; Hou, J.J.; Wang, X.; Yu, Y.; Li, H.J.; Li, Z.W.; Feng, Z.J.; Qu, H.; Wu, W.Y.; Guo, D.A. Venenum bufonis: An overview of its traditional use, natural product chemistry, pharmacology, pharmacokinetics and toxicology. J. Ethnopharmacol. 2019, 237, 215–235. [Google Scholar] [CrossRef]
- Zhang, D.M.; Liu, J.S.; Tang, M.K.; Yiu, A.; Cao, H.H.; Jiang, L.; Chan, J.Y.; Tian, H.Y.; Fung, K.P.; Ye, W.C. Bufotalin from Venenum Bufonis inhibits growth of multidrug resistant HepG2 cells through G2/M cell cycle arrest and apoptosis. Eur. J. Pharmacol. 2012, 692, 19–28. [Google Scholar] [CrossRef]
- Qiu, D.Z.; Zhang, Z.J.; Wu, W.Z.; Yang, Y.K. Bufalin, a component in Chansu, inhibits proliferation and invasion of hepatocellular carcinoma cells. BMC Complement. Altern. Med. 2013, 12, 185. [Google Scholar] [CrossRef] [Green Version]
- Qi, F.; Inagaki, Y.; Gao, B.; Cui, X.; Xu, H.; Kokudo, N.; Li, A.; Tang, W. Bufalin and cinobufagin induce apoptosis of human hepatocellular carcinoma cells via Fas- and mitochondria mediated pathways. Cancer Sci. 2011, 102, 951–958. [Google Scholar] [CrossRef]
- Zhang, L.; Liang, B.; Xu, H.; Gong, Y.; Hu, W.; Jin, Z.; Wu, X.; Chen, X.; Li, M.; Shi, L.; et al. Cinobufagin induces FOXO1-regulated apoptosis, proliferation, migration, and invasion by inhibiting G9a in non-small-cell lung cancer A549 cells. J. Ethnopharmacol. 2022, 291, 115095. [Google Scholar] [CrossRef]
- Ren, W.; Chen, S.; Liao, Y.; Li, S.; Ge, J.; Tao, F.; Huo, Q.; Zhang, Y.; Zhao, Z. Near-infrared fluorescent carbon dots encapsulated liposomes as multifunctional nano-carrier and tracer of the anticancer agent cinobufagin in vivo and in vitro. Colloids Surf. B Biointerfaces 2019, 174, 384–392. [Google Scholar] [CrossRef]
- Yang, A.L.; Wu, Q.; Hu, Z.D.; Wang, S.P.; Tao, Y.F.; Wang, A.M.; Sun, Y.X.; Li, X.L.; Dai, L.; Zhang, J. A network pharmacology approach to investigate the anticancer mechanism of cinobufagin against hepatocellular carcinoma via downregulation of EGFR-CDK2 signaling. Toxicol. Appl. Pharmacol. 2021, 431, 115739. [Google Scholar] [CrossRef]
- Kim, G.H.; Fang, X.Q.; Lim, W.J.; Park, J.; Kang, T.B.; Kim, J.H.; Lim, J.H. Cinobufagin Suppresses Melanoma Cell Growth by Inhibiting LEF1. Int. J. Mol. Sci. 2020, 21, 6706. [Google Scholar] [CrossRef]
- Ren, W.; Han, L.; Luo, M.; Bian, B.; Guan, M.; Yang, H.; Han, C.; Li, N.; Li, T.; Li, S.; et al. Multi-component identification and target cell-based screening of potential bioactive compounds in toad venom by UPLC coupled with high-resolution LTQ-Orbitrap MS and high-sensitivity Qtrap MS. Anal. Bioanal. Chem. 2018, 410, 4419–4435. [Google Scholar] [CrossRef]
- Zhang, J.; Hong, Y.; Xie, P.; Chen, Y.; Jiang, L.; Yang, Z.; Cao, G.; Chen, Z.; Liu, X.; Chen, Y.; et al. Spatial Lipidomics Reveals Anticancer Mechanisms of Bufalin in Combination with Cinobufagin in Tumor-Bearing Mice. Front. Pharmacol. 2020, 11, 593815. [Google Scholar] [CrossRef]
- Hou, Z.; Song, F.; Xing, J.; Zheng, Z.; Liu, S.; Liu, Z. Comprehensive fecal metabolomics and gut microbiota for the evaluation of the mechanism of Panax Ginseng in the treatment of Qi-deficiency liver cancer. J. Ethnopharmacol. 2022, 292, 115222. [Google Scholar] [CrossRef]
- Zhao, Y.Y.; Cheng, X.L.; Vaziri, N.D.; Liu, S.; Lin, R.C. UPLC-based metabonomic applications for discovering biomarkers of diseases in clinical chemistry. Clin. Biochem. 2014, 47, 16–26. [Google Scholar] [CrossRef] [Green Version]
- Tian, D.; Gao, Q.; Lin, J.; Chang, Z.; Wang, Y.; Shi, Y.; Su, R.; Han, Z.; Ma, D. Uncovering the mechanism of the Shenzhi Jiannao formula against vascular dementia using a combined network pharmacology approach and molecular biology. Phytomedicine 2021, 90, 153637. [Google Scholar] [CrossRef]
- Ploder, M.; Neurauter, G.; Spittler, A.; Schroecksnadel, K.; Roth, E.; Fuchs, D. Serum phenylalanine in patients post trauma and with sepsis correlate to neopterin concentrations. Amino Acids 2007, 35, 303–307. [Google Scholar] [CrossRef]
- Wiggins, T.; Kumar, S.; Markar, S.R.; Antonowicz, S.; Hanna, G.B. Tyrosine, phenylalanine, and tryptophan in gastroesophageal malignancy: A systematic review. Cancer Epidemiol. Biomarkers Prev. 2015, 24, 32–38. [Google Scholar] [CrossRef] [Green Version]
- Ali, M.R.; Wu, Y.; Han, T.; Zang, X.; Xiao, H.; Tang, Y.; Wu, R.; Fernandez, F.M.; El-Sayed, M.A. Simultaneous Time-Dependent Surface-Enhanced Raman Spectroscopy, Metabolomics, and Proteomics Reveal Cancer Cell Death Mechanisms Associated with Gold Nanorod Photothermal Therapy. J. Am. Chem. Soc. 2016, 138, 15434–15442. [Google Scholar] [CrossRef]
- Rohrig, F.; Schulze, A. The multifaceted roles of fatty acid synthesis in cancer. Nat. Rev. Cancer 2016, 16, 732–749. [Google Scholar] [CrossRef]
- Wu, H.; Cheng, H.; Luo, S.; Peng, C.; Zhou, A.; Chen, Z.; Wu, H.; Li, Q. Use of cellular metabolomics and lipidomics to decipher the mechanism of Huachansu injection-based intervention against human hepatocellular carcinoma cells. J. Pharm. Biomed. Anal. 2022, 212, 114654. [Google Scholar] [CrossRef]
- Crous-Maso, J.; Palomeras, S.; Relat, J.; Camo, C.; Martinez-Garza, U.; Planas, M.; Feliu, L.; Puig, T. (-)-Epigallocatechin 3-Gallate Synthetic Analogues Inhibit Fatty Acid Synthase and Show Anticancer Activity in Triple Negative Breast Cancer. Molecules 2018, 23, 1160. [Google Scholar] [CrossRef] [Green Version]
- Fan, H.; Huang, W.; Guo, Y.; Ma, X.; Yang, J. alpha-Linolenic Acid Suppresses Proliferation and Invasion in Osteosarcoma Cells via Inhibiting Fatty Acid Synthase. Molecules 2022, 27, 2741. [Google Scholar] [CrossRef]
- Kong, W.S.; Shen, F.X.; Xie, R.F.; Zhou, G.; Feng, Y.M.; Zhou, X. Bufothionine induces autophagy in H22 hepatoma-bearing mice by inhibiting JAK2/STAT3 pathway, a possible anti-cancer mechanism of cinobufacini. J. Ethnopharmacol. 2021, 270, 113848. [Google Scholar] [CrossRef]
- Chen, J.; Liu, Z.; Deng, F.; Liang, J.; Fan, B.; Zhen, X.; Tao, R.; Sun, L.; Zhang, S.; Cong, Z.; et al. Mechanisms of Lian-Gui-Ning-Xin-Tang in the treatment of arrhythmia: Integrated pharmacology and in vivo pharmacological assessment. Phytomedicine 2022, 99, 153989. [Google Scholar] [CrossRef] [PubMed]
- Li, F.J.; Hu, J.H.; Ren, X.; Zhou, C.M.; Liu, Q.; Zhang, Y.Q. Toad venom: A comprehensive review of chemical constituents, anticancer activities, and mechanisms. Arch. Pharm. 2021, 354, e2100060. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Lv, H.; Qiu, S.; Gao, L.; Ai, H. Plasma metabolic profiling and novel metabolite biomarkers for diagnosing prostate cancer. RSC Adv. 2017, 7, 30060–30069. [Google Scholar] [CrossRef] [Green Version]
- Lario, S.; Ramirez-Lazaro, M.J.; Sanjuan-Herraez, D.; Brunet-Vega, A.; Pericay, C.; Gombau, L.; Junquera, F.; Quintas, G.; Calvet, X. Plasma sample based analysis of gastric cancer progression using targeted metabolomics. Sci. Rep. 2017, 7, 17774. [Google Scholar] [CrossRef] [Green Version]
- Yao, X.; Li, Z.; Gong, X.; Fu, X.; Xiao, X.; He, M.; Huang, B.; Xu, Z. Total saponins extracted from Abrus cantoniensis Hance suppress hepatitis B virus replication in vitro and in rAAV8-1.3HBV transfected mice. J. Ethnopharmacol. 2020, 249, 112366. [Google Scholar] [CrossRef]
- Qi, G.; Wang, B.; Zhang, Y.; Li, H.; Li, C.; Xu, W.; Jin, Y. Living-Cell Imaging of Mitochondrial Membrane Potential Oscillation and Phenylalanine Metabolism Modulation during Periodic Electrostimulus. Anal. Chem. 2019, 91, 9571–9579. [Google Scholar] [CrossRef]
- Freund, A.; Laberge, R.M.; Demaria, M.; Campisi, J. Lamin B1 loss is a senescence-associated biomarker. Mol. Biol. Cell 2012, 23, 2066–2075. [Google Scholar] [CrossRef]
- Fang, Z.; Huang, H.; Wang, L.; Lin, Z. Identification of the alpha linolenic acid metabolism-related signature associated with prognosis and the immune microenvironment in nasopharyngeal carcinoma. Front. Endocrinol. 2022, 13, 968984. [Google Scholar] [CrossRef]
- Gu, Z.; Suburu, J.; Chen, H.; Chen, Y.Q. Mechanisms of omega-3 polyunsaturated fatty acids in prostate cancer prevention. Biomed. Res. Int. 2013, 2013, 824563. [Google Scholar] [CrossRef] [Green Version]
- Wang, S.C.; Sun, H.L.; Hsu, Y.H.; Liu, S.H.; Lii, C.K.; Tsai, C.H.; Liu, K.L.; Huang, C.S.; Li, C.C. alpha-Linolenic acid inhibits the migration of human triple-negative breast cancer cells by attenuating Twist1 expression and suppressing Twist1-mediated epithelial-mesenchymal transition. Biochem. Pharmacol. 2020, 180, 114152. [Google Scholar] [CrossRef]
- Li, Q.; Liu, X.; Wang, X.; Qiu, S.; Byambasuren, K.; Dang, L.; Wang, Z. Antiproliferative Ability and Fluorescence Tracking of alpha-Linolenic Acid-Loaded Microemulsion as Label-Free Delivery Carriers in MDA-MB-231 Cells. J. Agric. Food Chem. 2019, 67, 11518–11526. [Google Scholar] [CrossRef]
- Wiggins, A.K.; Mason, J.K.; Thompson, L.U. Growth and gene expression differ over time in alpha-linolenic acid treated breast cancer cells. Exp. Cell Res. 2015, 333, 147–154. [Google Scholar] [CrossRef]
- Kim, K.B.; Nam, Y.A.; Kim, H.S.; Hayes, A.W.; Lee, B.M. alpha-Linolenic acid: Nutraceutical, pharmacological and toxicological evaluation. Food Chem. Toxicol. 2014, 70, 163–178. [Google Scholar] [CrossRef]
- Mason, J.K.; Klaire, S.; Kharotia, S.; Wiggins, A.K.; Thompson, L.U. alpha-linolenic acid and docosahexaenoic acid, alone and combined with trastuzumab, reduce HER2-overexpressing breast cancer cell growth but differentially regulate HER2 signaling pathways. Lipids Health Dis. 2015, 14, 91. [Google Scholar] [CrossRef] [Green Version]
- Feng, S.; Xie, X.; Chen, C.; Zuo, S.; Zhao, X.; Li, H. Alpha-linolenic acid inhibits hepatocellular carcinoma cell growth through Farnesoid X receptor/beta-catenin signaling pathway. Nutr. Metab. 2022, 19, 57. [Google Scholar] [CrossRef]
- Lu, G.; Zhang, G.; Zheng, X.; Zeng, Y.; Xu, Z.; Zeng, W.; Wang, K. c9, t11- conjugated linoleic acid induces HCC cell apoptosis and correlation with PPAR-γ signaling pathway. Am. J. Transl. Res. 2015, 7, 2752–2763. [Google Scholar] [PubMed]
- Vecchini, A.; Ceccarelli, V.; Susta, F.; Caligiana, P.; Orvietani, P.; Binaglia, L.; Nocentini, G.; Riccardi, C.; Calviello, G.; Palozza, P.; et al. Dietary alpha-linolenic acid reduces COX-2 expression and induces apoptosis of hepatoma cells. J. Lipid Res. 2004, 45, 308–316. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhu, Q.; Zai, H.; Zhang, K.; Zhang, X.; Luo, N.; Li, X.; Hu, Y.; Wu, Y. L-norvaline affects the proliferation of breast cancer cells based on the microbiome and metabolome analysis. J. Appl. Microbiol. 2022, 133, 1014–1026. [Google Scholar] [CrossRef]
- Coker, O.O.; Liu, C.; Wu, W.K.K.; Wong, S.H.; Jia, W.; Sung, J.J.Y.; Yu, J. Altered gut metabolites and microbiota interactions are implicated in colorectal carcinogenesis and can be non-invasive diagnostic biomarkers. Microbiome 2022, 10, 35. [Google Scholar] [CrossRef]
- Qu, J.; Ke, F.; Liu, Z.; Yang, X.; Li, X.; Xu, H.; Li, Q.; Bi, K. Uncovering the mechanisms of dandelion against triple-negative breast cancer using a combined network pharmacology, molecular pharmacology and metabolomics approach. Phytomedicine 2022, 99, 153986. [Google Scholar] [CrossRef] [PubMed]
- Wei, M.M.; Wu, P.; Gao, J.N.; Wang, Y.J.; Zhou, J.L.; Xie, T.; Dong, X.M.; Fang, C.; Song, G.Q.; Huang, Z.H. A combination index and glycoproteomics-based approach revealed synergistic anticancer effects of curcuminoids of turmeric against prostate cancer PC3 cells. J. Ethnopharmacol. 2021, 267, 113467. [Google Scholar] [CrossRef] [PubMed]










| No | Compound | Type | RT (Min) | Chemical Formula | Error (Ppm) | [M+H]+ Measured Mass | [M+H]+ Calculated Mass |
|---|---|---|---|---|---|---|---|
| 1 | Bufotenidine-5-O- glucuronide | M | 2.54 | C19H27O7N2 | −0.399 | 395.18112 | 395.1812 |
| 2 | Bufotenidine | P | 2.62 | C13H19ON2 | −0.020 | 219.14917 | 219.1492 |
| 3 | Pitneloyl arginine | P | 5.28 | C13H25O5N4 | −1.060 | 317.18185 | 317.1820 |
| 4 | Suberoyl arginine | P | 7.43 | C14H27O5N4 | −0.533 | 331.19702 | 331.1976 |
| 5 | 1-hydroxylarenobufagin | M | 9.15 | C24H33O7 | −1.546 | 433.22128 | 433.2148 |
| 6 | 5-hydroxylarenobufagin | M | 9.65 | C24H33O7 | −1.477 | 433.22131 | 433.2148 |
| 7 | 16-hydroxylarenobufagin | M | 10.01 | C24H33O7 | −1.200 | 433.22153 | 433.2148 |
| 8 | Hydratalbufarenogin | M | 10.06 | C24H35O7 | −0.490 | 435.23709 | 435.2377 |
| 9 | 11α-hydroxytelocinobufagin | P | 10.52 | C24H35O6 | −1.491 | 419.24234 | 419.2428 |
| 10 | Ψ-Bufarenogin | P | 10.75 | C24H33O6 | −1.522 | 417.22638 | 417.2272 |
| 11 | Gamabufotalin | P | 11.34 | C24H35O5 | −1.837 | 403.24756 | 403.2479 |
| 12 | Bufarenogin | P | 11.94 | C24H33O6 | 0.539 | 417.22726 | 417.2272 |
| 13 | 19-hydroxylbufalin | P | 12.43 | C24H35O5 | −0.101 | 403.24799 | 403.2479 |
| 14 | Arenobufagin | P | 12.88 | C24H33O6 | −1.522 | 417.22733 | 417.2272 |
| 15 | Desacetylbufotalin | P | 13.16 | C24H35O5 | −0.324 | 403.24774 | 403.2479 |
| 16 | Hellebrigenol | P | 13.38 | C24H35O6 | 0.035 | 419.24289 | 419.2428 |
| 17 | Hellebrigenin | P | 13.76 | C24H33O6 | −0.132 | 417.22711 | 417.2272 |
| 18 | Bufotalinin | P | 15.97 | C24H31O6 | −1.241 | 415.21106 | 415.2115 |
| 19 | Telocinobufagin | P | 22.47 | C24H35O5 | −0.101 | 403.24774 | 403.2479 |
| 20 | Cinobufaginol | P | 24.43 | C26H35O7 | −0.784 | 459.23801 | 459.2377 |
| 21 | Desacetylcinobufagin | P | 26.84 | C24H33O5 | −0.901 | 401.23135 | 401.2323 |
| 22 | Bufalin | P | 26.95 | C24H35O4 | −1.178 | 387.25244 | 387.2530 |
| 23 | Resibufogenin | P | 28.69 | C24H33O4 | −0.950 | 385.23697 | 385.2373 |
| 24 | Cinobufagin | P | 28.89 | C26H35O6 | −0.265 | 443.24225 | 443.2428 |
| 25 | Unknown | M | 29.76 | C17H27O2 | −1.013 | 263.20032 | 263.2084 |
| 26 | Cinobufagin-3-O- glucuronide | M | 31.19 | C31H41O12 | −0.076 | 605.45453 | 605.4545 |
| 27 | Unknown | M | 31.57 | C20H33O2 | −1.824 | 305.24695 | 305.2553 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liang, P.; Ma, Y.; Yang, L.; Mao, L.; Sun, Q.; Sun, C.; Liu, Z.; Mazhar, M.; Yang, S.; Ren, W. Uncovering the Mechanisms of Active Components from Toad Venom against Hepatocellular Carcinoma Using Untargeted Metabolomics. Molecules 2022, 27, 7758. https://doi.org/10.3390/molecules27227758
Liang P, Ma Y, Yang L, Mao L, Sun Q, Sun C, Liu Z, Mazhar M, Yang S, Ren W. Uncovering the Mechanisms of Active Components from Toad Venom against Hepatocellular Carcinoma Using Untargeted Metabolomics. Molecules. 2022; 27(22):7758. https://doi.org/10.3390/molecules27227758
Chicago/Turabian StyleLiang, Pan, Yining Ma, Luyin Yang, Linshen Mao, Qin Sun, Changzhen Sun, Zengjin Liu, Maryam Mazhar, Sijin Yang, and Wei Ren. 2022. "Uncovering the Mechanisms of Active Components from Toad Venom against Hepatocellular Carcinoma Using Untargeted Metabolomics" Molecules 27, no. 22: 7758. https://doi.org/10.3390/molecules27227758
APA StyleLiang, P., Ma, Y., Yang, L., Mao, L., Sun, Q., Sun, C., Liu, Z., Mazhar, M., Yang, S., & Ren, W. (2022). Uncovering the Mechanisms of Active Components from Toad Venom against Hepatocellular Carcinoma Using Untargeted Metabolomics. Molecules, 27(22), 7758. https://doi.org/10.3390/molecules27227758