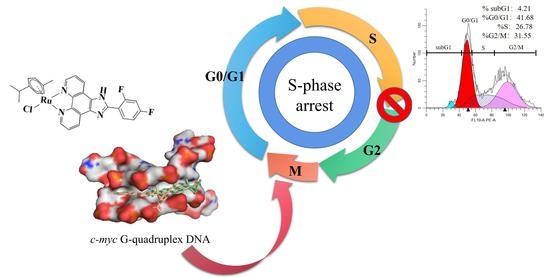
Arene Ru(II) Complexes with Difluorinated Ligands Act as Potential Inducers of S-Phase Arrest via the Stabilization of c-myc G-Quadruplex DNA
Abstract
:1. Introduction
2. Results and Discussion
2.1. Synthesis and Characterizations
2.2. G4 DNA Binding Behaviors
2.2.1. Molecular Docking
2.2.2. UV–Vis Spectra
2.2.3. Fluorescence Intercalator Displacement (FID) Assay
2.2.4. Fluorescence Resonance Energy Transfer (FRET) Melting Assay
2.3. Biological Studies
2.4. Joint Action of S-Phase Arrest and Apoptosis
3. Experimental Section
3.1. Materials and Methods
3.1.1. Chemicals
3.1.2. Instruments
3.1.3. Cell Lines and Culture
3.1.4. MTT Assay
3.1.5. n-Octanol-Water Partition Coefficient (logPo/w)
3.1.6. UV–Vis Titration
3.1.7. FID Assay
3.1.8. FRET Melting and Competitive FRET Assays
3.1.9. Molecular Docking
3.1.10. Flow Cytometric Analysis
3.2. Synthesis
3.2.1. Synthesis of 2-(2,3-Di-fluorophenyl) Imidazole [4,5f][1,10]-Phenanthroline (L1)
3.2.2. Synthesis of 2-(2,4-Di-fluorophenyl) Imidazole[4,5f][1,10]-Phenanthroline (L2)
3.2.3. Synthesis of [(η6-Benzene)Ru(L1)Cl]Cl (1)
3.2.4. Synthesis of [(η6-Toluene)Ru(L1)Cl]Cl (2)
3.2.5. Synthesis of [(η6-p-Cymene)Ru(L1)Cl]Cl (3)
3.2.6. Synthesis of [(η6-Benzene)Ru(L2)Cl]Cl (4)
3.2.7. Synthesis of [(η6-Toluene)Ru(L2)Cl]Cl (5)
3.2.8. Synthesis of [(η6-p-Cymene)Ru(L2)Cl]Cl (6)
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Sample Availability
References
- Chen, B.-J.; Wu, Y.-L.; Tanaka, Y.; Zhang, W. Small Molecules Targeting c-myc Oncogene: Promising Anti-Cancer Therapeutics. Int. J. Biol. Sci. 2014, 10, 1084–1096. [Google Scholar] [CrossRef] [PubMed]
- Brooks, T.A.; Kendrick, S.; Hurley, L. Making sense of G-quadruplex and i-motif functions in oncogene promoters. FEBS J. 2010, 277, 3459–3469. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Carabet, L.A.; Rennie, P.S.; Cherkasov, A. Therapeutic Inhibition of Myc in Cancer. Structural Bases and Computer-Aided Drug Discovery Approaches. Int. J. Mol. Sci. 2018, 20, 120. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chan, D.S.; Yang, H.; Kwan, M.H.; Cheng, Z.; Lee, P.; Bai, L.P.; Jiang, Z.H.; Wong, C.Y.; Fong, W.F.; Leung, C.H.; et al. Structure-based optimization of FDA-approved drug methylene blue as a c-myc G-quadruplex DNA stabilizer. Biochimie 2011, 93, 1055–1064. [Google Scholar] [CrossRef] [PubMed]
- Tan, C.P.; Zhong, Y.M.; Ji, L.N.; Mao, Z.W. Phosphorescent metal complexes as theranostic anticancer agents: Combining imaging and therapy in a single molecule. Chem. Sci. 2021, 12, 2357–2367. [Google Scholar] [CrossRef]
- Dyson, B.C.S.A.a.P.J. Ruthenium in Medicine Current Clinical Uses and Future Prospects. Platin. Met. Res. 2001, 45, 62–69. [Google Scholar]
- Vock, C.A.; Scolaro, C.; Phillips, R.D.; Scopelliti, R.; Sava, G.; Dyson, P.J. Synthesis, Characterization, and in Vitro Evaluation of Novel Ruthenium(II) η6-Arene Imidazole Complexes. J. Med. Chem. 2006, 49, 5552–5561. [Google Scholar] [CrossRef]
- Pavlovic, M.; Nikolic, S.; Gligorijevic, N.; Dojcinovic, B.; Arandelovic, S.; Grguric-Sipka, S.; Radulovic, S. New organoruthenium compounds with pyrido[2′,3′:5,6]pyrazino[2,3-f][1,10]phenanthroline: Synthesis, characterization, cytotoxicity, and investigation of mechanism of action. J. Biol. Inorg. Chem. 2019, 24, 297–310. [Google Scholar] [CrossRef]
- Yan, Y.K.; Melchart, M.; Habtemariam, A.; Sadler, P.J. Organometallic chemistry, biology and medicine: Ruthenium arene anticancer complexes. Chem. Commun. 2005, 38, 4764–4776. [Google Scholar] [CrossRef]
- Astudillo, D.; Silva, A.G.; Sanguinetti, M.E.; Villena, J.; Thomet, F.A. Cytotoxic organometallic [Ru(η6-anethole)(en)(X)]PF6 (X = Br or I) complexes: Synthesis, characterization and in vitro biological evaluation. Inorg. Chem. Commun. 2017, 84, 221–224. [Google Scholar] [CrossRef]
- Savi, A.; Gligorijevi, N.; Aranelovi, S.; Dojinovi, B.; Hecke, K.V.J.J.o.I.B. Antitumor activity of organoruthenium complexes with chelate aromatic ligands, derived from 1,10-phenantroline: Synthesis and biological activity. J. Inorg. Biochem. 2019, 202, 110869. [Google Scholar] [CrossRef] [PubMed]
- Fan, C.; Wu, Q.; Chen, T.; Zhang, Y.; Zheng, W.; Wang, Q.; Mei, W. Arene ruthenium(ii) complexes induce S-phase arrest in MG-63 cells through stabilization of c-myc G-quadruplex DNA. MedChemComm 2014, 5, 597–602. [Google Scholar] [CrossRef]
- Wu, Q.; Zheng, K.; Liao, S.; Ding, Y.; Li, Y.; Mei, W. Arene Ruthenium(II) Complexes as Low-Toxicity Inhibitor against the Proliferation, Migration, and Invasion of MDA-MB-231 Cells through Binding and Stabilizing c-myc G-Quadruplex DNA. Organometallics 2016, 35, 317–326. [Google Scholar] [CrossRef]
- Chen, Y.; Wu, Q.; Wang, X.; Xie, Q.; Tang, Y.; Lan, Y.; Zhang, S.; Mei, W. Microwave-Assisted Synthesis of Arene Ru(II) Complexes Induce Tumor Cell Apoptosis Through Selectively Binding and Stabilizing bcl-2 G-Quadruplex DNA. Materials 2016, 9, 386. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, Q.; Chen, T.; Zhao, Z.; Liao, S.; Zheng, W.J.D.T. Microwave-assisted synthesis of arene ruthenium(ii) complexes [(η6-RC6H5)Ru(m-MOPIP)Cl]Cl (R = -H and -CH3) as groove binder to c-myc G4 DNA. Dalton Trans. 2014, 43, 9216–9225. [Google Scholar] [CrossRef] [PubMed]
- Mei, W.-J.; Wu, Q.; Qian, J.-Y.; Peng, W.-H.; Shu, J. Organometallic compounds of ruthenium and their application in chemotherapy. In Organometallic Chemistry; The Royal Society of Chemistry: London, UK, 2021; Volume 43, pp. 35–62. [Google Scholar]
- Kolá?, M.H.; Tabarrini, O.J.J.o.M.C. Halogen Bonding in Nucleic Acid Complexes. J. Med. Chem. 2017, 60, 8681–8690. [Google Scholar] [CrossRef] [PubMed]
- Gupta, R.K.; Kumar, A.; Paitandi, R.P.; Singh, R.S.; Mukhopadhyay, S.; Verma, S.P.; Das, P.; Pandey, D.S. Heteroleptic arene Ru(II) dipyrrinato complexes: DNA, protein binding and anti-cancer activity against the ACHN cancer cell line. Dalton Trans. 2016, 45, 7163–7177. [Google Scholar] [CrossRef]
- Wumaier, M.; Shi, J.J.; Yao, T.M.; Hu, X.C.; Gao, R.R.; Shi, S. G-quadruplex and duplex DNA binding studies of novel Ruthenium(II) complexes containing ascididemin ligands. J. Inorg. Biochem. 2019, 196, 110681. [Google Scholar] [CrossRef]
- Monchaud, D.; Allain, C.; Teulade-Fichou, M.-P. Development of a fluorescent intercalator displacement assay (G4-FID) for establishing quadruplex-DNA affinity and selectivity of putative ligands. Bioorg. Med. Chem. Lett. 2006, 16, 4842–4845. [Google Scholar] [CrossRef]
- Monchaud, D.; Allain, C.; Teulade-Fichou, M.P. Thiazole orange: A useful probe for fluorescence sensing of G-quadruplex-ligand interactions. Nucleosides Nucleotides Nucleic Acids 2007, 26, 1585–1588. [Google Scholar] [CrossRef]
- Kaulage, M.H.; Maji, B.; Pasadi, S.; Ali, A.; Muniyappa, K.J.E.J.O.M.C. Targeting G-quadruplex DNA structures in the telomere and oncogene promoter regions by benzimidazole–carbazole ligands. Eur. J. Med. Chem. 2018, 148, 178–194. [Google Scholar] [CrossRef] [PubMed]
- Abbas, A.; Ali, B.; Kanwal; Khan, K.M.; Iqbal, J.; Ur Rahman, S.; Zaib, S.; Perveen, S. Synthesis and in vitro urease inhibitory activity of benzohydrazide derivatives, in silico and kinetic studies. Bioorg. Chem. 2019, 82, 163–177. [Google Scholar] [CrossRef] [PubMed]
- Choi, M.J.; No, E.S.; Thorat, D.A.; Jang, J.W.; Yang, H.; Lee, J.; Choo, H.; Kim, S.J.; Lee, C.S.; Ko, S.Y.; et al. Synthesis and biological evaluation of aryloxazole derivatives as antimitotic and vascular-disrupting agents for cancer therapy. J. Med. Chem. 2013, 56, 9008–9018. [Google Scholar] [CrossRef] [PubMed]
- Carella, A.; Roviello, V.; Iannitti, R.; Palumbo, R.; La Manna, S.; Marasco, D.; Trifuoggi, M.; Diana, R.; Roviello, G.N. Evaluating the biological properties of synthetic 4-nitrophenyl functionalized benzofuran derivatives with telomeric DNA binding and antiproliferative activities. Int. J. Biol. Macromol. 2019, 121, 77–88. [Google Scholar] [CrossRef] [PubMed]
- Nsmva, B.; Jcb, A.; Lpnr, A.; Amb, C.; Jmma, A.; Abp, A.J.C. Human cytotoxicity and octanol/water partition coefficients of fluorinated ionic liquids—ScienceDirect. Chemosphere 2019, 216, 576–586. [Google Scholar]
- Sarkar, B.; Mondal, A.; Madaan, Y.; Roy, N.; Moorthy, A.; Kuo, Y.-C.; Paira, P. Luminescent anticancer ruthenium(ii)-p-cymene complexes of extended imidazophenanthroline ligands: Synthesis, structure, reactivity, biomolecular interactions and live cell imaging. Dalton Trans. 2019, 48, 12257–12271. [Google Scholar] [CrossRef] [PubMed]
- Pany, S.P.; Bommisetti, P.; Diveshkumar, K.V.; Pradeepkumar, P.I. Benzothiazole hydrazones of furylbenzamides preferentially stabilize c-myc and c-KIT1 promoter G-quadruplex DNAs. Org. Biomol. Chem. 2016, 14, 5779–5793. [Google Scholar] [CrossRef]
- Liao, S.; Zhang, Z.; Wu, Q.; Wang, X.; Mei, W. Microwave-assisted synthesis of phenanthroimidazole derivatives as stabilizer of c-myc G-quadruplex DNA. Bioorg. Med. Chem. 2014, 22, 6503–6508. [Google Scholar] [CrossRef]









| Com. | DC50 (μM) | |||||
|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | |
| c-myc | 6.6 | 9.93 | 10.26 | 7.6 | 10.8 | 15.6 |
| Comp. | IC50/μM | ||||
|---|---|---|---|---|---|
| 570 nm | MCF-7 | A549 | HepG2 | U87 | LogPo/w |
| L1 | 4.25 ± 0.18 | 7.99 ± 0.05 | 0.29 ± 0.01 | 14.3 ± 0.47 | 0.375 |
| L2 | 1.48 ± 0.09 | 3.42 ± 0.13 | 0.12 ± 0.01 | 5.87 ± 0.27 | 0.218 |
| 1 | >100 | 95.03 ± 2.44 | >100 | >100 | −1.145 |
| 2 | >100 | 92.58 ± 5.34 | >100 | >100 | −1.251 |
| 3 | >100 | 41.58 ± 1.80 | >100 | 51.61 ± 1.64 | −0.6615 |
| 4 | >100 | 81.35 ± 4.96 | >100 | >100 | −1.414 |
| 5 | >100 | 80.47 ± 2.56 | >100 | >100 | −1.428 |
| 6 | >100 | 73.20 ± 0.63 | >100 | 23.75 ± 0.61 | −0.8077 |
| Cisplatin | 15.27 ± 0.18 | 17.27 ± 0.26 | 9.90 ± 0.06 | 32.59 ± 0.85 | - |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zeng, L.; Yuan, C.; Shu, J.; Qian, J.; Wu, Q.; Chen, Y.; Wu, R.; Ouyang, X.; Li, Y.; Mei, W. Arene Ru(II) Complexes with Difluorinated Ligands Act as Potential Inducers of S-Phase Arrest via the Stabilization of c-myc G-Quadruplex DNA. Molecules 2022, 27, 1897. https://doi.org/10.3390/molecules27061897
Zeng L, Yuan C, Shu J, Qian J, Wu Q, Chen Y, Wu R, Ouyang X, Li Y, Mei W. Arene Ru(II) Complexes with Difluorinated Ligands Act as Potential Inducers of S-Phase Arrest via the Stabilization of c-myc G-Quadruplex DNA. Molecules. 2022; 27(6):1897. https://doi.org/10.3390/molecules27061897
Chicago/Turabian StyleZeng, Liang, Chanling Yuan, Jing Shu, Jiayi Qian, Qiong Wu, Yanhua Chen, Ruzhen Wu, Xiaoming Ouyang, Yuan Li, and Wenjie Mei. 2022. "Arene Ru(II) Complexes with Difluorinated Ligands Act as Potential Inducers of S-Phase Arrest via the Stabilization of c-myc G-Quadruplex DNA" Molecules 27, no. 6: 1897. https://doi.org/10.3390/molecules27061897
APA StyleZeng, L., Yuan, C., Shu, J., Qian, J., Wu, Q., Chen, Y., Wu, R., Ouyang, X., Li, Y., & Mei, W. (2022). Arene Ru(II) Complexes with Difluorinated Ligands Act as Potential Inducers of S-Phase Arrest via the Stabilization of c-myc G-Quadruplex DNA. Molecules, 27(6), 1897. https://doi.org/10.3390/molecules27061897