Coffee Silverskin Phytocompounds as a Novel Anti-Aging Functional Food: A Pharmacoinformatic Approach Combined with In Vitro Study
Abstract
:1. Introduction
2. Results
2.1. Metabolites Profile of Two Coffee Silverskin Extract (CSE)
2.2. Pharmacoinformatics via Molecular Docking Simulation of Observed Compounds in Coffee Silverskin Extract
2.3. Antioxidant Capabilities of Two Coffee Silverskin Extracts
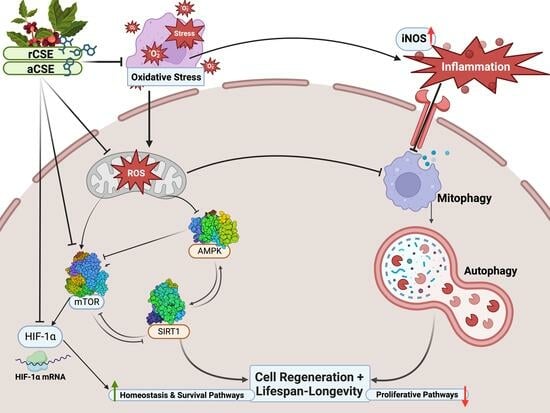
2.4. In Vitro Modulation of mTOR/AMPK/SIRT1 by Coffee Silverskin Extract
3. Discussion
4. Material and Methods
4.1. Chemical and Instrument
4.2. Preparation of CS Extract (CSE) via Ultrasound-Assisted Extraction (UAE) Method
4.3. Analyzing Untargeted Metabolomic Data Using HPLC-HRMS/ESI-MS
4.4. Pharmacoinformatic Approach via Molecular Docking Simulations
4.5. Antioxidants Activities Assay via DPPH and ABTS Inhibition
4.6. In Vitro Assay of mTOR/AMPK/SIRT1 Expressions
4.7. Data Analysis and Management
5. Conclusions
6. Patents
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Wong, Q.Y.A.; Chew, F.T. Defining skin aging and its risk factors: A systematic review and meta-analysis. Sci. Rep. 2021, 11, 22075. [Google Scholar] [CrossRef] [PubMed]
- Gunawan, W.B.; Abadi, M.N.P.; Fadhillah, F.S.; Nurkolis, F.; Pramono, A. The interlink between climate changes, gut microbiota, and aging processes. Hum. Nutr. Metab. 2023, 32, 200193. [Google Scholar] [CrossRef]
- Ge, Y.; Zhou, M.; Chen, C.; Wu, X.; Wang, X. Role of AMPK mediated pathways in autophagy and aging. Biochimie 2022, 195, 100–113. [Google Scholar] [CrossRef] [PubMed]
- Nurkolis, F.; Purnomo, A.F.; Alisaputra, D.; Gunawan, W.B.; Qhabibi, F.R.; Park, W.; Moon, M.; Taslim, N.A.; Park, M.N.; Kim, B. In silico and in vitro studies reveal a synergistic potential source of novel anti-ageing from two Indonesian green algae. J. Funct. Foods 2023, 104, 105555. [Google Scholar] [CrossRef]
- Singh, J.P.; Singh, B.; Kaur, A. Nutraceuticals and Functional Foods in Aging and Aging-Associated Diseases. In Nutrition, Food and Diet in Ageing and Longevity; Rattan, S.I.S., Kaur, G., Eds.; Springer International Publishing: Cham, Switzerland, 2021; pp. 221–238. ISBN 978-3-030-83017-5. [Google Scholar]
- Binns, C.W.; Lee, M.K.; Lee, A.H. Problems and Prospects: Public Health Regulation of Dietary Supplements. Annu. Rev. Public Health 2018, 39, 403–420. [Google Scholar] [CrossRef]
- Singh, S.; Garg, G.; Rizvi, S.I. Chapter 9—Plant polyphenols in balancing the redox state during aging. In Drug Discovery Update; Pandey, K.B., Suttajit, M., Eds.; Elsevier: Amsterdam, The Netherlands, 2023; pp. 181–195. ISBN 978-0-323-90581-7. [Google Scholar]
- Xu, Q.; Torres, J.E.; Hakim, M.; Babiak, P.M.; Pal, P.; Battistoni, C.M.; Nguyen, M.; Panitch, A.; Solorio, L.; Liu, J.C. Collagen- and hyaluronic acid-based hydrogels and their biomedical applications. Mater. Sci. Eng. R Rep. 2021, 146, 100641. [Google Scholar] [CrossRef]
- Mussatto, S.I.; Machado, E.M.S.; Martins, S.; Teixeira, J.A. Production, Composition, and Application of Coffee and Its Industrial Residues. Food Bioprocess Technol. 2011, 4, 661–672. [Google Scholar] [CrossRef]
- Narita, Y.; Inouye, K. Review on utilization and composition of coffee silverskin. Food Res. Int. 2014, 61, 16–22. [Google Scholar] [CrossRef]
- Iriondo-DeHond, A.; Martorell, P.; Genovés, S.; Ramón, D.; Stamatakis, K.; Fresno, M.; Molina, A.; Del Castillo, M.D. Coffee Silverskin Extract Protects against Accelerated Aging Caused by Oxidative Agents. Molecules 2016, 21, 721. [Google Scholar] [CrossRef]
- Fernandez-Gomez, B.; Lezama, A.; Amigo-Benavent, M.; Ullate, M.; Herrero, M.; Martín, M.Á.; Mesa, M.D.; del Castillo, M.D. Insights on the health benefits of the bioactive compounds of coffee silverskin extract. J. Funct. Foods 2016, 25, 197–207. [Google Scholar] [CrossRef]
- Iriondo-DeHond, A.; Herrera, T.; Del Castillo, M.D. Health Benefits of Silverskin. In Food Wastes and By-Products; John Wiley & Sons: Hoboken, NJ, USA, 2020; pp. 353–371. ISBN 9781119534167. [Google Scholar]
- Procentese, A.; Raganati, F.; Olivieri, G.; Russo, M.E.; Marzocchella, A. Combined antioxidant-biofuel production from coffee silverskin. Appl. Microbiol. Biotechnol. 2019, 103, 1021–1029. [Google Scholar] [CrossRef]
- Malara, A.; Paone, E.; Frontera, P.; Bonaccorsi, L.; Panzera, G.; Mauriello, F. Sustainable Exploitation of Coffee Silverskin in Water Remediation. Sustainability 2018, 10, 3547. [Google Scholar] [CrossRef]
- Nzekoue, F.K.; Angeloni, S.; Navarini, L.; Angeloni, C.; Freschi, M.; Hrelia, S.; Vitali, L.A.; Sagratini, G.; Vittori, S.; Caprioli, G. Coffee silverskin extracts: Quantification of 30 bioactive compounds by a new HPLC-MS/MS method and evaluation of their antioxidant and antibacterial activities. Food Res. Int. 2020, 133, 109128. [Google Scholar] [CrossRef] [PubMed]
- Mussatto, S.I. Chapter 11—Generating Biomedical Polyphenolic Compounds from Spent Coffee or Silverskin. In Coffee in Health and Disease Prevention; Preedy, V.R., Ed.; Academic Press: San Diego, CA, USA, 2015; pp. 93–106. ISBN 978-0-12-409517-5. [Google Scholar]
- del Pozo, C.; Rego, F.; Yang, Y.; Puy, N.; Bartrolí, J.; Fàbregas, E.; Bridgwater, A.V. Converting coffee silverskin to value-added products by a slow pyrolysis-based biorefinery process. Fuel Process. Technol. 2021, 214, 106708. [Google Scholar] [CrossRef]
- Iriondo-DeHond, A.; Rios, M.B.; Herrera, T.; Rodriguez-Bertos, A.; Nuñez, F.; San Andres, M.I.; Sanchez-Fortun, S.; del Castillo, M.D. Coffee Silverskin Extract: Nutritional Value, Safety and Effect on Key Biological Functions. Nutrients 2019, 11, 2693. [Google Scholar] [CrossRef]
- Andrade, N.; Peixoto, J.A.B.; Oliveira, M.B.P.P.; Martel, F.; Alves, R.C. Can coffee silverskin be a useful tool to fight metabolic syndrome? Front. Nutr. 2022, 9, 966734. [Google Scholar] [CrossRef] [PubMed]
- Giovannini, L.; Bianchi, S. Role of nutraceutical SIRT1 modulators in AMPK and mTOR pathway: Evidence of a synergistic effect. Nutrition 2017, 34, 82–96. [Google Scholar] [CrossRef]
- Kim, M.-J.; Ryu, G.R.; Kang, J.-H.; Sim, S.S.; Min, D.S.; Rhie, D.-J.; Yoon, S.H.; Hahn, S.J.; Jeong, I.-K.; Hong, K.-J.; et al. Inhibitory effects of epicatechin on interleukin-1β-induced inducible nitric oxide synthase expression in RINm5F cells and rat pancreatic islets by down-regulation of NF-κB activation. Biochem. Pharmacol. 2004, 68, 1775–1785. [Google Scholar] [CrossRef]
- Almaguer, G.; Ortiz-Vilchis, P.; Cordero, P.; Martinez-Vega, R.; Perez-Durán, J.; Meaney, E.; Villarreal, F.; Ceballos, G.; Nájera, N. Anticancer potential of (-)-epicatechin in a triple-negative mammary gland model. J. Pharm. Pharmacol. 2021, 73, 1675–1682. [Google Scholar] [CrossRef]
- Chang, S.-Y.; Ko, Y.; Kim, M.-J. Regulatory mechanisms of kaempferol on iNOS expression in RINm5F β-cells under exposure to interleukin-1β. Heliyon 2023, 9, e14818. [Google Scholar] [CrossRef]
- Kim, J.M.; Lee, E.K.; Kim, D.H.; Yu, B.P.; Chung, H.Y. Kaempferol modulates pro-inflammatory NF-κB activation by suppressing advanced glycation endproducts-induced NADPH oxidase. Age 2010, 32, 197–208. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Q.; Qu, Y.; Zhou, X.-G.; Chen, J.-N.; Luo, H.-R.; Wu, G.-S. A Dihydroflavonoid Naringin Extends the Lifespan of C. elegans and Delays the Progression of Aging-Related Diseases in PD/AD Models via DAF-16. Oxid. Med. Cell. Longev. 2020, 2020, 6069354. [Google Scholar] [CrossRef] [PubMed]
- Cui, Z.; Zhao, X.; Amevor, F.K.; Du, X.; Wang, Y.; Li, D.; Shu, G.; Tian, Y.; Zhao, X. Therapeutic application of quercetin in aging-related diseases: SIRT1 as a potential mechanism. Front. Immunol. 2022, 13, 943321. [Google Scholar] [CrossRef]
- Hid, E.J.; Mosele, J.I.; Prince, P.D.; Fraga, C.G.; Galleano, M. (−)-Epicatechin and cardiometabolic risk factors: A focus on potential mechanisms of action. Pflügers Arch.-Eur. J. Physiol. 2022, 474, 99–115. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.-D.; Kim, Y.-P.; Li, X.-C.; Baerson, S.R.; Agarwal, A.K.; Hodges, T.W.; Ferreira, D.; Nagle, D.G. Hypoxia-Inducible Factor-1 Activation by (−)-Epicatechin Gallate: Potential Adverse Effects of Cancer Chemoprevention with High-Dose Green Tea Extracts. J. Nat. Prod. 2004, 67, 2063–2069. [Google Scholar] [CrossRef] [PubMed]
- Alique, M.; Sánchez-López, E.; Bodega, G.; Giannarelli, C.; Carracedo, J.; Ramírez, R. Hypoxia-Inducible Factor-1α: The Master Regulator of Endothelial Cell Senescence in Vascular Aging. Cells 2020, 9, 195. [Google Scholar] [CrossRef]
- McGettrick, A.F.; O’Neill, L.A.J. The Role of HIF in Immunity and Inflammation. Cell Metab. 2020, 32, 524–536. [Google Scholar] [CrossRef]
- Aroufai, İ.A.; Sabuncu, M.; Dülger Altiner, D.; Sahan, Y. Antioxidant properties and bioaccessibility of coffee beans and their coffee silverskin grown in different countries. J. Food Meas. Charact. 2022, 16, 1873–1888. [Google Scholar] [CrossRef]
- Aissani, F.; Grara, N.; Bensouici, C.; Bousbia, A.; Ayed, H.; Idris, M.H.M.; Teh, L.K. Algerian Sonchus oleraceus L.: A comparison of different extraction solvent on phytochemical composition, antioxidant properties and anti-cholinesterase activity. Adv. Tradit. Med. 2022, 22, 383–394. [Google Scholar] [CrossRef]
- Bessada, S.M.F.; Alves, R.C.; Costa, A.S.G.; Nunes, M.A.; Oliveira, M.B.P.P. Coffea canephora silverskin from different geographical origins: A comparative study. Sci. Total Environ. 2018, 645, 1021–1028. [Google Scholar] [CrossRef] [PubMed]
- Papadopoli, D.; Boulay, K.; Kazak, L.; Pollak, M.; Mallette, F.A.; Topisirovic, I.; Hulea, L. mTOR as a central regulator of lifespan and aging. F1000Research 2019, 8, 998. [Google Scholar] [CrossRef] [PubMed]
- Chrienova, Z.; Nepovimova, E.; Kuca, K. The role of mTOR in age-related diseases. J. Enzym. Inhib. Med. Chem. 2021, 36, 1679–1693. [Google Scholar] [CrossRef] [PubMed]
- Steinberg, G.R.; Hardie, D.G. New insights into activation and function of the AMPK. Nat. Rev. Mol. Cell Biol. 2023, 24, 255–272. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Zhou, M.; Ge, Y.; Wang, X. SIRT1 and aging related signaling pathways. Mech. Ageing Dev. 2020, 187, 111215. [Google Scholar] [CrossRef] [PubMed]
- Steinberg, G.R.; Carling, D. AMP-activated protein kinase: The current landscape for drug development. Nat. Rev. Drug Discov. 2019, 18, 527–551. [Google Scholar] [CrossRef] [PubMed]
- Ren, Z.; He, H.; Zuo, Z.; Xu, Z.; Wei, Z.; Deng, J. The role of different SIRT1-mediated signaling pathways in toxic injury. Cell. Mol. Biol. Lett. 2019, 24, 36. [Google Scholar] [CrossRef]
- Sadria, M.; Layton, A.T. Interactions among mTORC, AMPK and SIRT: A computational model for cell energy balance and metabolism. Cell Commun. Signal. 2021, 19, 57. [Google Scholar] [CrossRef]
- Cetrullo, S.; D’Adamo, S.; Tantini, B.; Borzi, R.M.; Flamigni, F. mTOR, AMPK, and Sirt1: Key Players in Metabolic Stress Management. Crit. Rev. Eukaryot. Gene Expr. 2015, 25, 59–75. [Google Scholar] [CrossRef]
- Hardinsyah, H.; Gunawan, W.B.; Nurkolis, F.; Alisaputra, D.; Kurniawan, R.; Mayulu, N.; Taslim, N.A.; Tallei, T.E. Antiobesity potential of major metabolites from Clitoria ternatea kombucha: Untargeted metabolomic profiling and molecular docking simulations. Curr. Res. Food Sci. 2023, 6, 100464. [Google Scholar] [CrossRef]
- Permatasari, H.K.; Nurkolis, F.; Hardinsyah, H.; Taslim, N.A.; Sabrina, N.; Ibrahim, F.M.; Visnu, J.; Kumalawati, D.A.; Febriana, S.A.; Sudargo, T.; et al. Metabolomic Assay, Computational Screening, and Pharmacological Evaluation of Caulerpa racemosa as an Anti-obesity With Anti-aging by Altering Lipid Profile and Peroxisome Proliferator-Activated Receptor-γ Coactivator 1-α Levels. Front. Nutr. 2022, 9, 939073. [Google Scholar] [CrossRef]
- Permatasari, H.K.; Nurkolis, F.; Gunawan, W.B.; Yusuf, V.M.; Yusuf, M.; Kusuma, R.J.; Sabrina, N.; Muharram, F.R.; Taslim, N.A.; Mayulu, N.; et al. Modulation of gut microbiota and markers of metabolic syndrome in mice on cholesterol and fat enriched diet by butterfly pea flower kombucha. Curr. Res. Food Sci. 2022, 5, 1251–1265. [Google Scholar] [CrossRef] [PubMed]
- Kurniawan, R.; Nurkolis, F.; Taslim, N.A.; Subali, D.; Surya, R.; Gunawan, W.B.; Alisaputra, D.; Mayulu, N.; Salindeho, N.; Kim, B. Carotenoids Composition of Green Algae Caulerpa racemosa and Their Antidiabetic, Anti-Obesity, Antioxidant, and Anti-Inflammatory Properties. Molecules 2023, 28, 3267. [Google Scholar] [CrossRef] [PubMed]




| Sample | Observed Compounds | Molecular Formula | RT (Min) | Observed MW (m/z) | PubChem ID or Substance ID | CAS Number | Type |
|---|---|---|---|---|---|---|---|
| aCSE | Epicatechin | C15H14O6 | 16.07 | 289.8855 | 72276 | 490-46-0 | Flavonoids |
| Kaempferol | C15H10O6 | 10.14 | 285.9800 | 5280863 | 520-18-3 | Flavonoids | |
| Quercitrin | C21H20O11 | 9.02 | 448.1100 | 5280459 | 522-12-3 | Flavonoids | |
| 4-Hydroxycinnamic acid | C9H8O3 | 19.55 | 164.5500 | 637542 | 501-98-4 | Polyphenols (Phenolic Acids) | |
| Gallic acid | C7H6O5 | 16.02 | 170.1400 | 370 | 149-91-7 | Polyphenols (Phenolic Acids) | |
| rCSE | Shikimic Acid | C7H10O5 | 10.90 | 174.0855 | 8742 | 138-59-0 | Polyphenols (Phenolic Acids) |
| (2R,3S,4S,5R,6S)-2-(hydroxymethyl)-6-[7-hydroxy-3-[(2S,3R,4S,5S,6R)-3,4,5-trihydroxy-6-(hydroxymethyl)oxan-2-yl]oxy-2-(3,4,5-trihydroxyphenyl)chromenylium-5-yl]oxyoxane-3,4,5-triol | C27H31O17+ | 12.92 | 627.1522 | 10100906 | 17670-06-3 | Flavonoids | |
| Caffeic acid | C9H8O4 | 9.10 | 179.8800 | 689043 | 331-39-5 | Polyphenols (Phenolic Acids) | |
| Naringin | C27H32O14 | 15.13 | 580.0300 | 442428 | 10236-47-2 | Flavonoids | |
| Rutin | C27H30O16 | 13.56 | 610.0225 | 5280805 | 153-18-4 | Flavonoids | |
| (+)-Catechin | C15H14O6 | 14.30 | 290.0100 | 9064 | 154-23-4 | Flavonoids |
| No. | Target Proteins or Receptors | PDB ID | Docking Site (x;y;z) | Docking Area (x;y;z) | RMSD (Å) | ΔG (kcal/mol) | Number in Cluster (/100) | Judgment (<2 Å) |
|---|---|---|---|---|---|---|---|---|
| 1 | iNOS | 3E7G | 55.022, 21.817, 78.677 | 40 × 40 × 40 | 1.789 | −6.67 | 98 | Valid |
| 2 | mTOR | 3FAP | −9.233, 26.776, 35.832 | 46 × 40 × 42 | 1.422 | −21.75 | 100 | Valid |
| 3 | ROS1 Kinase | 3ZBF | 42.521, 19.649, 3.987 | 40 × 40 × 40 | 1.216 | −7.83 | 90 | Valid |
| 4 | HIF-1α | 1H2N | 19.984, 25.64, 28.282 | 40 × 40 × 40 | 1.128 | −3.82 | 100 | Valid |
| No. | Substance | Number in Cluster (/100) | dG (kcal/mol) | Ki | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 3E7G | 3FAP | 3ZBF | 1H2N | 3E7G | 3FAP | 3ZBF | 1H2N | 3E7G | 3FAP | 3ZBF | 1H2N | ||
| Control | |||||||||||||
| 1 | S-ibuprofen | 33 | −4.73 | 128.28 uM | |||||||||
| 2 | Quercetin | 29 | −5.96 | 41.26 uM | |||||||||
| 3 | Luteolin | 96 | −6.68 | 6.86 uM | |||||||||
| 4 | Genistein | 49 | −6.77 | 9.14 uM | |||||||||
| aCSE | |||||||||||||
| 1 | Epicatechin | 92 | 27 | 68 | 61 | −6.15 | −6.07 | −6.10 | −7.00 | 15.85 uM | 20.92 uM | 14.20 uM | 5.59 uM |
| 2 | Gallic acid | 39 | 67 | 87 | 100 | −4.03 | −3.95 | −3.72 | −4.97 | 710.63 uM | 617.81 uM | 1.11 mM | 86.21 uM |
| 3 | 4-Hydroxycinnamic acid | 55 | 64 | 98 | 99 | −4.37 | −4.46 | −4.95 | −5.20 | 433.26 uM | 838.56 uM | 180.35 uM | 133.46 uM |
| 4 | Kaempferol | 31 | 41 | 100 | 48 | −5.86 | −6.17 | −6.45 | −6.25 | 39.82 uM | 17.09 uM | 17.88 uM | 24.17 uM |
| 5 | Quercitrin | 30 | 24 | 29 | 36 | −5.41 | −7.10 | −6.57 | −6.07 | 20.98 uM | 2.86 uM | 5.08 uM | 1.63 uM |
| rCSE | |||||||||||||
| 1 | Caffeic acid | 26 | 74 | 96 | 75 | −4.19 | −4.54 | −5.16 | −5.13 | 490.27 uM | 303.08 uM | 114.81 uM | 117.47 uM |
| 2 | (+)-Catechin | 51 | 29 | 43 | 59 | −6.07 | −6.48 | −6.15 | −6.28 | 33.94 uM | 16.65 uM | 17.30 uM | 19.97 uM |
| 3 | (2R,3S,4S,5R,6S)-2-(hydroxymethyl)-6-[7-hydroxy-3-[(2S,3R,4S,5S,6R)-3,4,5-trihydroxy-6-(hydroxymethyl)oxan-2-yl]oxy-2-(3,4,5-trihydroxyphenyl)chromenylium-5-yl]oxyoxane-3,4,5-triol | 37 | 9 | 33 | 28 | −3.98 | −5.54 | −4.41 | −3.39 | 36.27 uM | 19.12 uM | 7.57 uM | 3.26 mM |
| 4 | Naringin | 24 | 16 | 56 | 61 | −5.78 | −8.37 | −6.47 | −6.37 | 3.58 uM | 82.91 nM | 2.59 uM | 21.26 uM |
| 5 | Rutin | 12 | 8 | 18 | 18 | −3.79 | −5.93 | −4.98 | −3.38 | 119.48 uM | 12.61 uM | 5.66 uM | 3.35 mM |
| 6 | Shikimic Acid | 64 | 35 | 82 | 90 | −4.05 | −4.22 | −3.33 | −4.99 | 525.14 uM | 398.89 uM | 2.12 mM | 128.97 uM |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hayes, C.; Nurkolis, F.; Laksemi, D.A.A.S.; Chung, S.; Park, M.N.; Choi, M.; Choi, J.; Darmaputra, I.G.N.; Gunawan, W.B.; Lele, J.A.J.M.N.; et al. Coffee Silverskin Phytocompounds as a Novel Anti-Aging Functional Food: A Pharmacoinformatic Approach Combined with In Vitro Study. Molecules 2023, 28, 7037. https://doi.org/10.3390/molecules28207037
Hayes C, Nurkolis F, Laksemi DAAS, Chung S, Park MN, Choi M, Choi J, Darmaputra IGN, Gunawan WB, Lele JAJMN, et al. Coffee Silverskin Phytocompounds as a Novel Anti-Aging Functional Food: A Pharmacoinformatic Approach Combined with In Vitro Study. Molecules. 2023; 28(20):7037. https://doi.org/10.3390/molecules28207037
Chicago/Turabian StyleHayes, Clarin, Fahrul Nurkolis, Dewa Ayu Agus Sri Laksemi, Sanghyun Chung, Moon Nyeo Park, Min Choi, Jinwon Choi, I Gusti Nyoman Darmaputra, William Ben Gunawan, Juan Alessandro Jeremis Maruli Nura Lele, and et al. 2023. "Coffee Silverskin Phytocompounds as a Novel Anti-Aging Functional Food: A Pharmacoinformatic Approach Combined with In Vitro Study" Molecules 28, no. 20: 7037. https://doi.org/10.3390/molecules28207037
APA StyleHayes, C., Nurkolis, F., Laksemi, D. A. A. S., Chung, S., Park, M. N., Choi, M., Choi, J., Darmaputra, I. G. N., Gunawan, W. B., Lele, J. A. J. M. N., Khumaidi, M. A., Taslim, N. A., & Kim, B. (2023). Coffee Silverskin Phytocompounds as a Novel Anti-Aging Functional Food: A Pharmacoinformatic Approach Combined with In Vitro Study. Molecules, 28(20), 7037. https://doi.org/10.3390/molecules28207037