Associations of NR5A2 Gene Polymorphisms with the Clinicopathological Characteristics and Survival of Gastric Cancer
Abstract
:1. Introduction
2. Results
2.1. Associations between Clinicopathological Variables and Overall Survival
| Variable | Patients, n = 944 | MST (Months) | Log-Rank p | HR (95% CI) d |
|---|---|---|---|---|
| Age (years) | ||||
| ≤60 | 442 | 88 | 0.239 | 1.000 |
| >60 | 502 | 59 | 1.118 (0.927–1.349) | |
| Sex | ||||
| Male | 727 | 70 | 0.536 | 1.000 |
| Female | 217 | 63 | 1.071 (0.860–1.335) | |
| Tumor Size | ||||
| ≤5 cm | 581 | 98 | <0.001 | 1.000 |
| >5 cm | 363 | 48 | 1.422 (1.178–1.716) | |
| Location | ||||
| Non-Cardia | 626 | 78 | 0.253 | 1.000 |
| Cardia | 318 | 63 | 0.891 (0.730–1.088) | |
| Histological Types a | ||||
| Intestinal | 370 | 74 | 0.436 | 1.000 |
| Diffuse | 497 | 60 | 1.069 (0.918–1.244) | |
| Differentiation a | ||||
| Well to Moderate | 305 | 80 | 0.518 | 1.000 |
| Poorly | 493 | 59 | 1.165 (0.942–1.443) | |
| Mucinous/Signet-Ring Cell | 69 | 62 | 1.199 (0.825–1.742) | |
| Lauren a | ||||
| 1 | 399 | 76 | <0.001 | 1.000 |
| 2 | 541 | 50 | 1.467 (1.211–1.776) | |
| Depth of Invasion b | ||||
| T1 | 182 | 84 | <0.001 | 1.000 |
| T2 | 138 | 78 | 0.540 (0.408–0.714) | |
| T3 | 8 | 70 | 0.800 (0.608–1.054) | |
| T4 | 594 | 51 | 1.008 (0.416–2.442) | |
| Lymph Node Metastasis c | ||||
| N0 | 378 | 81 | <0.001 | 1.000 |
| N1/N2/N3 | 566 | 43 | 1.814 (1.480–2.222) | |
| Distant Metastasis | ||||
| M0 | 886 | 74 | 0.004 | 1.000 |
| M1 | 58 | 26 | 1.646 (1.165–2.326) | |
| TNM Stage | ||||
| I | 250 | 83 | <0.001 | 1.000 |
| II | 203 | 88 | 1.231 (0.909–1.666) | |
| III | 458 | 41 | 1.949 (1.524–2.492) | |
| IV | 25 | 44 | 2.046 (1.163–3.600) | |
| Chemotherapy | ||||
| No | 638 | 61 | 0.684 | 1.000 |
| Yes | 306 | 69 | 1.043 (0.852–1.276) |
2.2. Association Analyses of NR5A2 rs3790843 and rs3790844 Genotypes with Clinicopathological Features
| Variable | rs3790843 (n = 907) | rs3790844 (n = 912) | |||||
|---|---|---|---|---|---|---|---|
| TT | TC/CC | p | GG | GA/AA | p | ||
| Age (years) | ≤60 | 192 | 237 | 0.610 | 182 | 249 | 0.380 |
| >60 | 222 | 256 | 217 | 264 | |||
| Sex | Male | 321 | 375 | 0.601 | 311 | 391 | 0.539 |
| Female | 93 | 118 | 88 | 122 | |||
| Location | Non-Cardia Cancer | 279 | 320 | 0.432 | 264 | 338 | 0.930 |
| Cardia Cancer | 135 | 173 | 135 | 175 | |||
| Tumor Size | ≤5 cm | 261 | 302 | 0.581 | 243 | 324 | 0.486 |
| >5 cm | 153 | 191 | 156 | 189 | |||
| Lymph Node Metastasis a | N0 | 180 | 182 | 0.044 | 168 | 197 | 0.257 |
| N1/N2/N3 | 234 | 311 | 231 | 316 | |||
| Distant Metastasis | M0 | 398 | 456 | 0.020 | 382 | 478 | 0.098 |
| M1 | 16 | 37 | 9 | 35 | |||
| Histological Types b | Intestinal | 168 | 187 | 0.273 | 156 | 205 | 0.541 |
| Diffuse | 207 | 270 | 206 | 270 | |||
| Differentiation b | Well to Moderate | 137 | 155 | 0.556 | 127 | 170 | 0.687 |
| Poorly | 208 | 267 | 205 | 270 | |||
| Mucinous/Signet-Ring Cell | 30 | 35 | 30 | 35 | |||
| Lauren b | 1 | 182 | 199 | 0.253 | 169 | 218 | 0.450 |
| 2 | 229 | 293 | 227 | 294 | |||
| Chemotherapy | No | 275 | 333 | 0.721 | 266 | 348 | 0.709 |
| Yes | 139 | 160 | 133 | 165 | |||
| Depth of Invasion c | T1 | 86 | 93 | 0.854 | 76 | 101 | 0.929 |
| T2 | 60 | 69 | 59 | 71 | |||
| T3 | 3 | 3 | 2 | 4 | |||
| T4 | 257 | 320 | 254 | 326 | |||
| TNM Stage | I | 120 | 118 | 0.369 | 111 | 128 | 0.533 |
| II | 83 | 111 | 78 | 119 | |||
| III | 198 | 248 | 198 | 250 | |||
| IV | 10 | 13 | 9 | 13 | |||
| Haplotype | Frequencies | Patients/Deaths | HR (95% CI) a |
|---|---|---|---|
| TTGG | 0.415 | 379/184 | 1.00 |
| CTGA | 0.408 | 373/170 | 0.90 (0.73–1.10) |
| CCAA | 0.079 | 72/30 | 0.79 (0.54–1.17) |
| TTGA | 0.036 | 33//11 | 0.54 (0.30–1.00) |
| CTAA | 0.024 | 22//6 | 0.47 (0.21–1.07) |
| CTGG | 0.015 | 14//10 | 1.55 (0.81–2.94) |
2.3. Associations of NR5A2 rs3790843 and rs3790844 with Clinical Outcomes of OS
| Genetic Model | Genotypes | Patients | Deaths | MST (Months) | Log-Rank p | HR (95%CI) a |
|---|---|---|---|---|---|---|
| rs3790843 | ||||||
| Codominant Model | TT | 414 | 196 | 59 | 0.668 | 1.000 |
| CT | 410 | 187 | 70 | 0.934 (0.765–1.142) | ||
| CC | 83 | 36 | 88 | 0.872 (0.611–1.244) | ||
| Dominant Model | TT | 414 | 196 | 59 | 0.415 | 1.000 |
| CT or CC | 493 | 223 | 70 | 0.924 (0.762–1.119) | ||
| Recessive Model | TT or CT | 824 | 383 | 70 | 0.551 | 1.000 |
| CC | 83 | 36 | 88 | 0.902 (0.641–1.269) | ||
| rs3790844 | ||||||
| Codominant Model | GG | 399 | 197 | 52 | 0.075 | 1.000 |
| GA | 416 | 186 | 78 | 0.853 (0.698–1.043) | ||
| AA | 97 | 37 | 98 | 0.699 (0.492–0.993) | ||
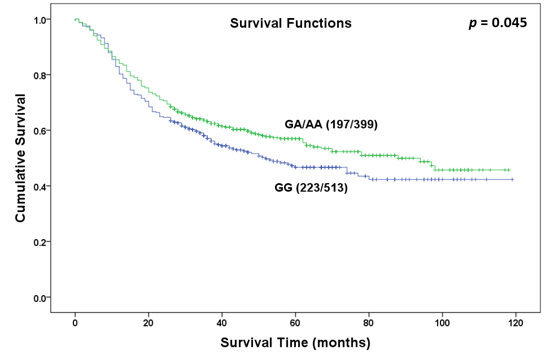
| Dominant Model | GG | 399 | 197 | 52 | 0.045 | 1.000 |
| GA or AA | 513 | 223 | 88 | 0.823 (0.679–0.997) | ||
| Recessive Model | GG or GA | 815 | 383 | 65 | 0.103 | 1.000 |
| AA | 97 | 37 | 98 | 0.757 (0.540–1.061) | ||
3. Discussion

| rs3790843 | rs3790844 | |||||||
|---|---|---|---|---|---|---|---|---|
| TT | TC/CC | HR (95% CI) e | p | GG | GA/AA | HR (95% CI) e | p | |
| Age (years) | ||||||||
| ≤60 | 88/192 | 108/237 | 0.977 (0.737–1.294) | 0.869 | 90/182 | 107/249 | 0.806 (0.609–1.067) | 0.132 |
| >60 | 108/222 | 115/256 | 0.873 (0.672–1.136) | 0.313 | 107/217 | 116/264 | 0.837 (0.644–1.089) | 0.185 |
| Sex | ||||||||
| Male | 144/321 | 175/375 | 1.044 (0.838–1.302) | 0.700 | 149/311 | 172/391 | 0.877 (0.705–1.093) | 0.243 |
| Female | 52/93 | 48/118 | 0.619 (0.417–0.916) | 0.017 | 48/88 | 51/122 | 0.660 (0.445–0.981) | 0.040 |
| Location | ||||||||
| Non–CardiaCancer | 133/279 | 148/320 | 0.948 (0.750–1.198) | 0.652 | 133/264 | 148/338 | 0.810 (0.641–1.024) | 0.077 |
| CardiaCancer | 63/135 | 75/173 | 0.861 (0.615–1.204) | 0.381 | 64/135 | 75/175 | 0.840 (0.601–1.174) | 0.308 |
| Tumor Size | ||||||||
| ≤5 cm | 108/261 | 127/302 | 1.044 (0.807–1.349) | 0.745 | 106/243 | 132/324 | 0.927 (0.718–1.197) | 0.559 |
| >5 cm | 88/153 | 96/191 | 0.747 (0.558–0.999) | 0.049 | 91/156 | 91/189 | 0.693 (0.517–0.928) | 0.014 |
| Lymph Node Metastasis a | ||||||||
| N0 | 64/180 | 60/182 | 0.947 (0.666–1.347) | 0.763 | 63/168 | 63/197 | 0.865 (0.610–1.226) | 0.414 |
| N1/N2/N3 | 132/234 | 163/311 | 0.829 (0.659–1.043) | 0.109 | 134/231 | 160/316 | 0.744 (0.591–0.937) | 0.012 |
| Distant Metastasis | ||||||||
| M0 | 188/398 | 200/456 | 0.886 (0.726–1.081) | 0.232 | 188/382 | 202/478 | 0.793 (0.650–0.968) | 0.022 |
| M1 | 8/16 | 23/37 | 1.340 (0.599–2.998) | 0.476 | 7/9 | 21/35 | 1.189 (0.544–2.599) | 0.664 |
| Histological Types b | ||||||||
| Intestinal | 71/168 | 80/187 | 1.041 (0.756–1.434) | 0.805 | 69/156 | 84/205 | 0.952 (0.692–1.309) | 0.762 |
| Diffuse | 108/207 | 127/270 | 0.826 (0.639–1.068) | 0.145 | 111/206 | 123/270 | 0.735 (0.568–0.950) | 0.019 |
| Differentiation b | ||||||||
| Well to Moderate | 57/137 | 67/155 | 1.099 (0.772–1.565) | 0.601 | 56/127 | 69/170 | 0.962 (0.676–1.368) | 0.828 |
| Poorly | 104/208 | 126/267 | 0.882 (0.680–1.144) | 0.344 | 105/205 | 125/270 | 0.822 (0.634–1.067) | 0.140 |
| Others c | 18/30 | 14/35 | 0.522 (0.259–1.056) | 0.07 | 19/30 | 13/35 | 0.397 (0.195–0.808) | 0.011 |
| Lauren b | ||||||||
| 1 | 69/182 | 78/199 | 1.049 (0.759–1.451) | 0.771 | 69/169 | 80/218 | 0.899 (0.652–1.241) | 0.655 |
| 2 | 125/229 | 144/293 | 0.824 (0.648–1.047) | 0.113 | 126/227 | 142/294 | 0.763 (0.600–0.970) | 0.027 |
| Chemotherapy | ||||||||
| No | 132/275 | 151/333 | 0.908 (0.719–1.147) | 0.416 | 136/266 | 149/348 | 0.767 (0.608–0.968) | 0.025 |
| Yes | 64/139 | 72/160 | 0.957 (0.683–1.340) | 0.799 | 61/133 | 74/165 | 0.953 (0.679–1.338) | 0.783 |
| Depth of Invasion d | ||||||||
| T1 | 25/86 | 34/93 | 1.297 (0.774–2.173) | 0.324 | 24/76 | 33/101 | 1.051 (0.621–1.779) | 0.852 |
| T2 | 31/60 | 24/69 | 0.669 (0.393–1.141) | 0.140 | 31/59 | 25/71 | 0.648 (0.382–1.099) | 0.107 |
| T3 | 1/3 | 2/3 | 1.405 (0.125–15.838) | 0.782 | 1/2 | 2/4 | 0.809 (0.071–9.157) | 0.864 |
| T4 | 134/257 | 159/320 | 0.880 (0.699–1.108) | 0.276 | 135/254 | 157/326 | 0.809 (0.642–1.019) | 0.072 |
| TNM Stage | ||||||||
| I | 40/120 | 39/118 | 0.992 (0.638–1.542) | 0.972 | 40/111 | 40/128 | 0.862 (0.556–1.336) | 0.506 |
| II | 38/83 | 38/111 | 0.743 (0.474–1.166) | 0.196 | 37/78 | 41/119 | 0.711 (0.456–1.109) | 0.132 |
| III | 111/198 | 135/248 | 0.861 (0.670–1.108) | 0.245 | 113/198 | 132/250 | 0.800 (0.622–1.029) | 0.082 |
| IV | 4/10 | 8/13 | 1.855 (0.557–6.171) | 0.314 | 4/9 | 7/13 | 1.257 (0.368–4.302) | 0.715 |
| Variables | β | SE | HR b | 95% CI | p Value |
|---|---|---|---|---|---|
| Age a | 0.067 | 0.100 | 1.070 | (0.878–1.302) | 0.503 |
| Sex | 0.04 | 0.118 | 1.041 | (0.825–1.313) | 0.743 |
| Lymph Node Metastasis | 0.567 | 0.112 | 1.763 | (1.414–2.198) | <0.001 |
| rs3790844 (GG vs. GA/AA) | −0.243 | 0.099 | 0.784 | (0.646–0.951) | 0.014 |
| rs3790843 (TT vs. CT/CC) | −0.152 | 0.101 | 0.859 | (0.705–1.046) | 0.131 |
4. Experimental Section
4.1. Study Subjects
4.2. Genotyping
4.3. Statistical Methods
5. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Jemal, A.; Bray, F.; Center, M.M.; Ferlay, J.; Ward, E.; Forman, D. Global cancer statistics. CA Cancer J. Clin. 2011, 61, 69–90. [Google Scholar] [CrossRef] [PubMed]
- Parkin, D.M.; Bray, F.; Ferlay, J.; Pisani, P. Global cancer statistics, 2002. CA Cancer J. Clin. 2005, 55, 74–108. [Google Scholar] [CrossRef]
- Santoro, R.; Carboni, F.; Lepiane, P.; Ettorre, G.M.; Santoro, E. Clinicopathological features and prognosis of gastric cancer in young european adults. Br. J. Surg. 2007, 94, 737–742. [Google Scholar] [CrossRef]
- Correa, P. Human gastric carcinogenesis: A multistep and multifactorial process—First american cancer society award lecture on cancer epidemiology and prevention. Cancer Res. 1992, 52, 6735–6740. [Google Scholar] [PubMed]
- Resende, C.; Ristimaki, A.; Machado, J.C. Genetic and epigenetic alteration in gastric carcinogenesis. Helicobacter 2010, 15 (S1), 34–39. [Google Scholar] [CrossRef]
- Ludwig, J.A.; Weinstein, J.N. Biomarkers in cancer staging, prognosis and treatment selection. Nat. Rev. Cancer 2005, 5, 845–856. [Google Scholar] [CrossRef] [PubMed]
- Panani, A.D. Cytogenetic and molecular aspects of gastric cancer: Clinical implications. Cancer Lett. 2008, 266, 99–115. [Google Scholar] [CrossRef] [PubMed]
- Becker, K.F.; Keller, G.; Hoefler, H. The use of molecular biology in diagnosis and prognosis of gastric cancer. Surg. Oncol. 2000, 9, 5–11. [Google Scholar] [CrossRef] [PubMed]
- Fayard, E.; Auwerx, J.; Schoonjans, K. LRH-1: An orphan nuclear receptor involved in development, metabolism and steroidogenesis. Trends Cell Biol. 2004, 14, 250–260. [Google Scholar] [CrossRef] [PubMed]
- Fernandez-Marcos, P.J.; Auwerx, J.; Schoonjans, K. Emerging actions of the nuclear receptor LRH-1 in the gut. Biochim. Biophys. Acta 2011, 1812, 947–955. [Google Scholar] [CrossRef] [PubMed]
- Venteclef, N.; Jakobsson, T.; Ehrlund, A.; Damdimopoulos, A.; Mikkonen, L.; Ellis, E.; Nilsson, L.M.; Parini, P.; Janne, O.A.; Gustafsson, J.A.; et al. GPS2-dependent corepressor/SUMO pathways govern anti-inflammatory actions of LRH-1 and LXRβ in the hepatic acute phase response. Genes Dev. 2010, 24, 381–395. [Google Scholar] [CrossRef] [PubMed]
- Clyne, C.D.; Kovacic, A.; Speed, C.J.; Zhou, J.; Pezzi, V.; Simpson, E.R. Regulation of aromatase expression by the nuclear receptor LRH-1 in adipose tissue. Mol. Cell. Endocrinol. 2004, 215, 39–44. [Google Scholar] [CrossRef] [PubMed]
- Clyne, C.D.; Speed, C.J.; Zhou, J.; Simpson, E.R. Liver receptor homologue-1 (LRH-1) regulates expression of aromatase in preadipocytes. J. Biol. Chem. 2002, 277, 20591–20597. [Google Scholar] [CrossRef] [PubMed]
- Lazarus, K.A.; Wijayakumara, D.; Chand, A.L.; Simpson, E.R.; Clyne, C.D. Therapeutic potential of liver receptor homolog-1 modulators. J. Steroid Biochem. Mol. Biol. 2012, 130, 138–146. [Google Scholar] [CrossRef] [PubMed]
- Benod, C.; Vinogradova, M.V.; Jouravel, N.; Kim, G.E.; Fletterick, R.J.; Sablin, E.P. Nuclear receptor liver receptor homologue 1 (LRH-1) regulates pancreatic cancer cell growth and proliferation. Proc. Natl. Acad. Sci. USA 2011, 108, 16927–16931. [Google Scholar] [CrossRef] [PubMed]
- Petersen, G.M.; Amundadottir, L.; Fuchs, C.S.; Kraft, P.; Stolzenberg-Solomon, R.Z.; Jacobs, K.B.; Arslan, A.A.; Bueno-de-Mesquita, H.B.; Gallinger, S.; Gross, M.; et al. A genome-wide association study identifies pancreatic cancer susceptibility loci on chromosomes 13q22.1, 1q32.1 and 5p15.33. Nat. Genet. 2010, 42, 224–228. [Google Scholar] [CrossRef] [PubMed]
- Schoonjans, K.; Dubuquoy, L.; Mebis, J.; Fayard, E.; Wendling, O.; Haby, C.; Geboes, K.; Auwerx, J. Liver receptor homolog 1 contributes to intestinal tumor formation through effects on cell cycle and inflammation. Proc. Natl. Acad. Sci. USA 2005, 102, 2058–2062. [Google Scholar] [CrossRef] [PubMed]
- Rausa, F.M.; Galarneau, L.; Belanger, L.; Costa, R.H. The nuclear receptor fetoprotein transcription factor is coexpressed with its target gene HNF-3β in the developing murine liver, intestine and pancreas. Mech. Dev. 1999, 89, 185–188. [Google Scholar] [CrossRef]
- Galarneau, L.; Pare, J.F.; Allard, D.; Hamel, D.; Levesque, L.; Tugwood, J.D.; Green, S.; Belanger, L. The α1-fetoprotein locus is activated by a nuclear receptor of the drosophila FTZ-F1 family. Mol. Cell. Biol. 1996, 16, 3853–3865. [Google Scholar] [PubMed]
- Botrugno, O.A.; Fayard, E.; Annicotte, J.S.; Haby, C.; Brennan, T.; Wendling, O.; Tanaka, T.; Kodama, T.; Thomas, W.; Auwerx, J.; et al. Synergy between LRH-1 and β-catenin induces G1 cyclin-mediated cell proliferation. Mol. Cell 2004, 15, 499–509. [Google Scholar] [CrossRef] [PubMed]
- Lew, D.J.; Dulic, V.; Reed, S.I. Isolation of three novel human cyclins by rescue of G1 cyclin (cln) function in yeast. Cell 1991, 66, 1197–1206. [Google Scholar] [CrossRef] [PubMed]
- Sherr, C.J. Mammalian G1 cyclins. Cell 1993, 73, 1059–1065. [Google Scholar] [CrossRef] [PubMed]
- Wimmel, A.; Lucibello, F.C.; Sewing, A.; Adolph, S.; Muller, R. Inducible acceleration of G1 progression through tetracycline-regulated expression of human cyclin E. Oncogene 1994, 9, 995–997. [Google Scholar] [PubMed]
- Florenes, V.A.; Faye, R.S.; Maelandsmo, G.M.; Nesland, J.M.; Holm, R. Levels of cyclin D1 and D3 in malignant melanoma: Deregulated cyclin D3 expression is associated with poor clinical outcome in superficial melanoma. Clin. Cancer Res. 2000, 6, 3614–3620. [Google Scholar] [PubMed]
- Richter, J.; Wagner, U.; Kononen, J.; Fijan, A.; Bruderer, J.; Schmid, U.; Ackermann, D.; Maurer, R.; Alund, G.; Knonagel, H.; et al. High-throughput tissue microarray analysis of cyclin E gene amplification and overexpression in urinary bladder cancer. Am. J. Pathol. 2000, 157, 787–794. [Google Scholar] [CrossRef] [PubMed]
- Kamai, T.; Takagi, K.; Asami, H.; Ito, Y.; Oshima, H.; Yoshida, K.I. Decreasing of p27kip1 and cyclin E protein levels is associated with progression from superficial into invasive bladder cancer. Br. J. Cancer 2001, 84, 1242–1251. [Google Scholar] [CrossRef] [PubMed]
- Heah, K.G.; Hassan, M.I.; Huat, S.C. p53 Expression as a marker of microinvasion in oral squamous cell carcinoma. Asian Pac. J. Cancer Prev. 2011, 12, 1017–1022. [Google Scholar] [PubMed]
- Ahn, M.J.; Kim, B.H.; Jang, S.J.; Hong, E.K.; Lee, W.M.; Baik, H.K.; Park, H.K.; Lee, C.B.; Ki, M. Expression of cyclin D1 and cyclin E in human gastric carcinoma and its clinicopathologic significance. J. Korean Med. Sci. 1998, 13, 513–518. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.L.; Zheng, D.Z.; Lan, F.H.; Deng, X.J.; Zeng, J.; Li, C.J.; Wang, R.; Zhu, Z.Y. Increased expression of hLRH-1 in human gastric cancer and its implication in tumorigenesis. Mol. Cell. Biochem. 2008, 308, 93–100. [Google Scholar] [CrossRef] [PubMed]
- He, T.C.; Sparks, A.B.; Rago, C.; Hermeking, H.; Zawel, L.; da Costa, L.T.; Morin, P.J.; Vogelstein, B.; Kinzler, K.W. Identification of c-myc as a target of the APC pathway. Science 1998, 281, 1509–1512. [Google Scholar] [CrossRef] [PubMed]
- Shtutman, M.; Zhurinsky, J.; Simcha, I.; Albanese, C.; D’Amico, M.; Pestell, R.; Ben-Ze’ev, A. The cyclin D1 gene is a target of the β-catenin/LEF-1 pathway. Proc. Natl. Acad. Sci. USA 1999, 96, 5522–5527. [Google Scholar] [CrossRef] [PubMed]
- Tetsu, O.; McCormick, F. β-catenin regulates expression of cyclin D1 in colon carcinoma cells. Nature 1999, 398, 422–426. [Google Scholar] [CrossRef] [PubMed]
- Kato, J.; Matsushime, H.; Hiebert, S.W.; Ewen, M.E.; Sherr, C.J. Direct binding of cyclin D to the retinoblastoma gene product (pRb) and pRb phosphorylation by the cyclin D-dependent kinase CKD4. Genes Dev. 1993, 7, 331–342. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.K.; Diehl, J.A. Nuclear cyclin D1: An oncogenic driver in human cancer. J. Cell. Physiol. 2009, 220, 292–296. [Google Scholar] [CrossRef] [PubMed]
- Annicotte, J.S.; Chavey, C.; Servant, N.; Teyssier, J.; Bardin, A.; Licznar, A.; Badia, E.; Pujol, P.; Vignon, F.; Maudelonde, T.; et al. The nuclear receptor liver receptor homolog-1 is an estrogen receptor target gene. Oncogene 2005, 24, 8167–8175. [Google Scholar] [PubMed]
- Thiruchelvam, P.T.; Lai, C.F.; Hua, H.; Thomas, R.S.; Hurtado, A.; Hudson, W.; Bayly, A.R.; Kyle, F.J.; Periyasamy, M.; Photiou, A.; et al. The liver receptor homolog-1 regulates estrogen receptor expression in breast cancer cells. Breast Cancer Res. Treat. 2011, 127, 385–396. [Google Scholar] [CrossRef] [PubMed]
- Miki, Y.; Clyne, C.D.; Suzuki, T.; Moriya, T.; Shibuya, R.; Nakamura, Y.; Ishida, T.; Yabuki, N.; Kitada, K.; Hayashi, S.; et al. Immunolocalization of liver receptor homologue-1 (LRH-1) in human breast carcinoma: Possible regulator of insitu steroidogenesis. Cancer Lett. 2006, 244, 24–33. [Google Scholar] [CrossRef] [PubMed]
- Chand, A.L.; Herridge, K.A.; Thompson, E.W.; Clyne, C.D. The orphan nuclear receptor LRH-1 promotes breast cancer motility and invasion. Endocr. Relat. Cancer 2010, 17, 965–975. [Google Scholar] [CrossRef] [PubMed]
- Lai, C.F.; Flach, K.D.; Alexi, X.; Fox, S.P.; Ottaviani, S.; Thiruchelvam, P.T.; Kyle, F.J.; Thomas, R.S.; Launchbury, R.; Hua, H.; et al. Co-regulated gene expression by oestrogen receptor alpha and liver receptor homolog-1 is a feature of the oestrogen response in breast cancer cells. Nucleic Acids Res. 2013, 41, 10228–10240. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Bai, J.; Tan, Y.; Wang, S.; Tian, Y.; Gong, W.; Zhou, Y.; Gao, Y.; Zhou, J.; Zhang, Z. Genetic variant in PSCA predicts survival of diffuse-type gastric cancer in a Chinese population. Int. J. Cancer 2011, 129, 1207–1213. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Wang, M.; Gu, D.; Wu, D.; Zhang, X.; Gong, W.; Tan, Y.; Zhou, J.; Wu, X.; Tang, C.; et al. Association of XRCC1 gene polymorphisms with the survival and clinicopathological characteristics of gastric cancer. DNA Cell Biol. 2013, 32, 111–118. [Google Scholar] [CrossRef] [PubMed]
© 2014 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, X.; Gu, D.; Du, M.; Wang, M.; Cao, C.; Shen, L.; Kuang, M.; Tan, Y.; Huo, X.; Gong, W.; et al. Associations of NR5A2 Gene Polymorphisms with the Clinicopathological Characteristics and Survival of Gastric Cancer. Int. J. Mol. Sci. 2014, 15, 22902-22917. https://doi.org/10.3390/ijms151222902
Zhang X, Gu D, Du M, Wang M, Cao C, Shen L, Kuang M, Tan Y, Huo X, Gong W, et al. Associations of NR5A2 Gene Polymorphisms with the Clinicopathological Characteristics and Survival of Gastric Cancer. International Journal of Molecular Sciences. 2014; 15(12):22902-22917. https://doi.org/10.3390/ijms151222902
Chicago/Turabian StyleZhang, Xunlei, Dongying Gu, Mulong Du, Meilin Wang, Chunxiang Cao, Lili Shen, Meng Kuang, Yongfei Tan, Xinying Huo, Weida Gong, and et al. 2014. "Associations of NR5A2 Gene Polymorphisms with the Clinicopathological Characteristics and Survival of Gastric Cancer" International Journal of Molecular Sciences 15, no. 12: 22902-22917. https://doi.org/10.3390/ijms151222902
APA StyleZhang, X., Gu, D., Du, M., Wang, M., Cao, C., Shen, L., Kuang, M., Tan, Y., Huo, X., Gong, W., Xu, Z., Chen, J., Zhang, Z., & Tang, C. (2014). Associations of NR5A2 Gene Polymorphisms with the Clinicopathological Characteristics and Survival of Gastric Cancer. International Journal of Molecular Sciences, 15(12), 22902-22917. https://doi.org/10.3390/ijms151222902