Biochemical Properties and Structure Analysis of a DAG-Like Lipase from Malassezia globosa
Abstract
:1. Introduction
2. Results and Discussion
2.1. Sequence Analysis of MgMDL2

2.2. Expression and Purification of MgMDL2

2.3. Biochemical Properties of MgMDL2
2.3.1. Effect of Temperature on Lipase Activity and Thermostability

2.3.2. Effect of pH on Lipase Activity and Stability

2.3.3. Effect of Chemicals and Organic Solvents on MgMDL2 Activity
| Reagents | Relative Activity (%) a | |
|---|---|---|
| 5 mM | 30% (v/v) | |
| Control | 100 | 100 |
| Ni2+ | 36 ± 1 | - |
| Zn2+ | 61 ± 4 | - |
| Mn2+ | 35 ± 4 | - |
| Mg2+ | 53 ± 4 | - |
| Fe2+ | 48 ± 2 | - |
| Ca2+ | 42 ± 1 | - |
| EDTA | 68 ± 1 | - |
| Methanol | - | 98 ± 2 |
| Ethanol | - | 97 ± 4 |
| Acetone | - | 85 ± 4 |
| Isopropanol | - | 41 ± 2 |
2.3.4. Substrate Specificity of MgMDL2

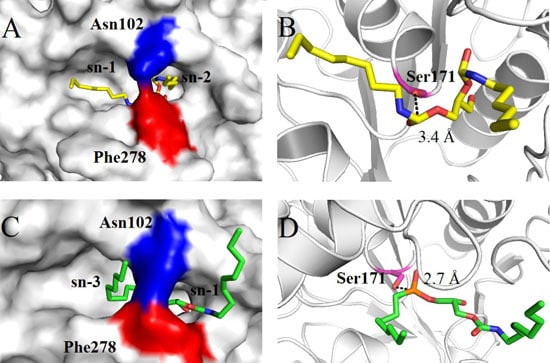
2.4. Molecular Basis for Substrate Selectivity of MgMDL2


3. Experimental Section
3.1. Strains, Plasmids, Chemicals and Materials
3.2. Vector Construction and Transformation of P. pastoris
3.3. Expression and Purification of Recombinant Enzyme
3.4. Biochemical Characterization of Recombinant Enzyme
3.4.1. Enzyme Activity Determination
3.4.2. Determining Temperature-Optimum of Activity and Thermostability of Lipase
3.4.3. Determining pH-Optimum of Activity and pH Stability of Lipase
3.4.4. Substrates Specificity
3.4.5. Effect of Metal Ions on the Enzyme Activity
3.4.6. Effect of Organic Solvents on the Enzyme Activity
3.4.7. Detection of Glycolysation Modification
3.5. Sequence and Structure Analysis
3.5.1. Sequence Analysis
3.5.2. Construction of the Structure in the Open Conformation
3.5.3. Construction of a Model in Complex with Substrate Analogue
3.5.4. MD Simulations
4. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Bisogno, T.; Howell, F.; Williams, G.; Minassi, A.; Cascio, M.G.; Ligresti, A.; Matias, I.; Schiano-Moriello, A.; Paul, P.; Williams, E.J.; et al. Cloning of the first sn1-DAG lipases points to the spatial and temporal regulation of endocannabinoid signaling in the brain. J. Cell Biol. 2003, 163, 463–468. [Google Scholar] [CrossRef] [PubMed]
- Kohnz, R.A.; Nomura, D.K. Chemical approaches to therapeutically target the metabolism and signaling of the endocannabinoid 2-AG and eicosanoids. Chem. Soc. Rev. 2014, 43, 6859–6869. [Google Scholar] [CrossRef] [PubMed]
- Xu, D.; Sun, L.J.; Chen, H.Y.; Lan, D.M.; Wang, Y.H.; Yang, B. Enzymatic synthesis of diacylglycerols enriched with conjugated linoleic acid by a novel lipase from Malassezia globosa. J. Am. Oil Chem. Soc. 2012, 89, 1259–1266. [Google Scholar]
- Wang, W.F.; Li, T.; Qin, X.L.; Ning, Z.X.; Yang, B.; Wang, Y.H. Production of lipase SMG1 and its application in synthesizing diacylglyecrol. J. Mol. Catal. B 2012, 77, 87–91. [Google Scholar] [CrossRef]
- Yamaguchi, S.; Takeuchi, K.; Mase, T.; Matsuura, A. Efficient expression of mono- and diacylglycerol lipase gene from Penicillium camembertii U-150 in Aspergillus oryzae under the control of its own promoter. Biosci. Biotechnol. Biochem. 1997, 61, 800–805. [Google Scholar] [CrossRef] [PubMed]
- Chahinian, H.; Vanot, G.; Ibrik, A.; Rugani, N.; Sarda, L.; Comeau, L.C. Production of extracellular lipases by Penicillium cyclopium purification and characterization of a partial acylglycerol lipase. Biosci. Biotechnol. Biochem. 2000, 64, 215–222. [Google Scholar] [CrossRef] [PubMed]
- Mase, T.; Matsumiya, Y.; Akiba, T. Purification and characterization of a new lipase from fusarium sp ym-30. Biosci. Biotechnol. Biochem. 1995, 59, 1771–1772. [Google Scholar] [CrossRef] [PubMed]
- Toida, J.; Kondoh, K.; Fukuzawa, M.; Ohnishi, K.; Sekiguchi, J. purification and characterization of a lipase from aspergillus-oryzae. Biosci. Biotechnol. Biochem. 1995, 59, 1199–1203. [Google Scholar] [CrossRef] [PubMed]
- Tsuchiya, A.; Nakazawa, H.; Toida, J.; Ohnishi, K.; Sekiguchi, J. Cloning and nucleotide sequence of the mono- and diacylglycerol lipase gene (mdlB) of Aspergillus oryzae. FEMS Microbiol. Lett. 1996, 143, 63–67. [Google Scholar] [CrossRef] [PubMed]
- DeAngelis, Y.M.; Saunders, C.W.; Johnstone, K.R.; Reeder, N.L.; Coleman, C.G.; Kaczvinsky, J.R., Jr.; Gale, C.; Walter, R.; Mekel, M.; Lacey, M.P.; et al. Isolation and expression of a Malassezia globosa lipase gene, LIP1. J. Investig. Dermatol. 2007, 127, 2138–2146. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Lan, D.M.; Wang, Q.; Gao, C.L.; Li, Z.G.; Yang, B.; Wang, Y.H. A “bridge-like” structure responsible for the substrate selectivity of mono- and diacylglycerol lipase from Aspergillus oryzae. J. Mol. Catal. B 2013, 97, 144–149. [Google Scholar] [CrossRef]
- Xu, T.; Liu, L.; Hou, S.; Xu, J.; Yang, B.; Wang, Y.; Liu, J. Crystal structure of a mono- and diacylglycerol lipase from Malassezia globosa reveals a novel lid conformation and insights into the substrate specificity. J. Struct. Biol. 2012, 178, 363–369. [Google Scholar] [CrossRef] [PubMed]
- Derewenda, U.; Swenson, L.; Green, R.; Wei, Y.; Dodson, G.G.; Yamaguchi, S.; Haas, M.J.; Derewenda, Z.S. An unusual buried polar cluster in a family of fungal lipases. Nat. Struct. Biol. 1994, 1, 36–47. [Google Scholar] [CrossRef] [PubMed]
- Derewenda, U.; Brzozowski, A.M.; Lawson, D.M.; Derewenda, Z.S. Catalysis at the interface: The anatomy of a conformational change in a triglyceride lipase. Biochemistry (Mosc.) 1992, 31, 1532–1541. [Google Scholar] [CrossRef]
- Derewenda, U.; Swenson, L.; Wei, Y.; Green, R.; Kobos, P.M.; Joerger, R.; Haas, M.J.; Derewenda, Z.S. Conformational lability of lipases observed in the absence of an oil-water interface: Crystallographic studies of enzymes from the fungi Humicola lanuginosa and Rhizopus delemar. J. Lipid Res. 1994, 35, 524–534. [Google Scholar] [PubMed]
- Skropeta, D. The effect of individual N-glycans on enzyme activity. Bioorg. Med. Chem. 2009, 17, 2645–2653. [Google Scholar] [CrossRef] [PubMed]
- Tan, Z.B.; Li, J.F.; Li, X.T.; Gu, Y.; Wu, M.C.; Wu, J.; Wang, J.Q. A unique mono- and diacylglycerol lipase from Penicillium cyclopium: Heterologous expression, biochemical characterization and molecular basis for its substrate selectivity. PLoS One 2014, 9, e102040. [Google Scholar] [CrossRef] [PubMed]
- Cereghino, J.L.; Cregg, J.M. Heterologous protein expression in the methylotrophic yeast Pichia pastoris. FEMS Microbiol. Rev. 2000, 24, 45–66. [Google Scholar] [CrossRef] [PubMed]
- Solá, R.J.; Griebenow, K. Effects of glycosylation on the stability of protein pharmaceuticals. J. Pharm. Sci. 2009, 98, 1223–1245. [Google Scholar] [CrossRef] [PubMed]
- Kavitha, M.; Shanthi, C. Isolation and Characterization of Cold active lipase producing Pseudomonas sp 4 from Marine samples of Tamilnadu Coast. Res. J. Biotechnol. 2013, 8, 57–62. [Google Scholar]
- Jeon, J.H.; Kim, J.T.; Kim, Y.J.; Kim, H.K.; Lee, H.S.; Kang, S.G.; Kim, S.J.; Lee, J.H. Cloning and characterization of a new cold-active lipase from a deep-sea sediment metagenome. Appl. Microbiol. Biotechnol. 2009, 81, 865–874. [Google Scholar] [CrossRef] [PubMed]
- Parra, L.P.; Reyes, F.; Acevedo, J.P.; Salazar, O.; Andrews, B.A.; Asenjo, J.A. Cloning and fusion expression of a cold-active lipase from marine Antarctic origin. Enzym. Microb. Technol. 2008, 42, 371–377. [Google Scholar] [CrossRef]
- Hårdeman, F.; Sjöling, S. Metagenomic approach for the isolation of a novel low-temperature-active lipase from uncultured bacteria of marine sediment. FEMS Microbiol. Ecol. 2007, 59, 524–534. [Google Scholar] [CrossRef] [PubMed]
- Ryu, H.; Kim, H.; Choi, W.; Kim, M.; Park, S.; Han, N.; Oh, T.; Lee, J. New cold-adapted lipase from Photobacterium lipolyticum sp. nov. that is closely related to filamentous fungal lipases. Appl. Microbiol. Biotechnol. 2006, 70, 321–326. [Google Scholar] [CrossRef] [PubMed]
- Sulong, M.R.; Zaliha Raja Abd Rahman, R.N.; Salleh, A.B.; Basri, M. A novel organic solvent tolerant lipase from Bacillus sphaericus 205y: Extracellular expression of a novel OST-lipase gene. Protein Expr. Purif. 2006, 49, 190–195. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ji, Q.; Xiao, S.; He, B.; Liu, X. Purification and characterization of an organic solvent-tolerant lipase from Pseudomonas aeruginosa LX1 and its application for biodiesel production. J. Mol. Catal. B 2010, 66, 264–269. [Google Scholar] [CrossRef]
- Liu, R.; Jiang, X.; Mou, H.; Guan, H.; Hwang, H.; Li, X. A novel low-temperature resistant alkaline lipase from a soda lake fungus strain Fusarium solani N4–2 for detergent formulation. Biochem. Eng. J. 2009, 46, 265–270. [Google Scholar] [CrossRef]
- Gao, C.; Lan, D.; Liu, L.; Zhang, H.; Yang, B.; Wang, Y. Site-directed mutagenesis studies of the aromatic residues at the active site of a lipase from Malassezia globosa. Biochimie 2014, 102, 29–36. [Google Scholar] [CrossRef] [PubMed]
- Kanwar, S.S.; Kaushal, R.K.; Jawed, A.; Gupta, R.; Chimni, S.S. Methods for inhibition of residual lipase activity in colorimetric assay: A comparative study. Indian J. Biochem. Biophys. 2005, 42, 233–237. [Google Scholar] [PubMed]
- Xu, Y.; Guo, S.H.; Wang, W.F.; Wang, Y.H.; Yang, B. Enzymatic hydrolysis of palm stearin to produce diacylglycerol with a highly thermostable lipase. Eur. J. Lipid Sci. Technol. 2013, 115, 564–570. [Google Scholar] [CrossRef]
- Boratyn, G.M.; Camacho, C.; Cooper, P.S.; Coulouris, G.; Fong, A.; Ma, N.; Madden, T.L.; Matten, W.T.; McGinnis, S.D.; Merezhuk, Y.; et al. BLAST: A more efficient report with usability improvements. Nucleic Acids Res. 2013, 41, W29–W33. [Google Scholar] [CrossRef] [PubMed]
- Petersen, T.N.; Brunak, S.; von Heijne, G.; Nielsen, H. SignalP 4.0: Discriminating signal peptides from transmembrane regions. Nat. Methods 2011, 8, 785–786. [Google Scholar] [CrossRef] [PubMed]
- Larkin, M.A.; Blackshields, G.; Brown, N.P.; Chenna, R.; McGettigan, P.A.; McWilliam, H.; Valentin, F.; Wallace, I.M.; Wilm, A.; Lopez, R.; et al. Clustal W and Clustal X version 2.0. Bioinformatics 2007, 23, 2947–2948. [Google Scholar] [CrossRef] [PubMed]
- Gouet, P.; Courcelle, E.; Stuart, D.I.; Metoz, F. ESPript: Analysis of multiple sequence alignments in PostScript. Bioinformatics 1999, 15, 305–308. [Google Scholar] [CrossRef] [PubMed]
- Nardini, M.; Lang, D.A.; Liebeton, K.; Jaeger, K.E.; Dijkstra, B.W. Crystal structure of pseudomonas aeruginosa lipase in the open conformation. The prototype for family I.1 of bacterial lipases. J. Biol. Chem. 2000, 275, 31219–31225. [Google Scholar] [CrossRef] [PubMed]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xu, H.; Lan, D.; Yang, B.; Wang, Y. Biochemical Properties and Structure Analysis of a DAG-Like Lipase from Malassezia globosa. Int. J. Mol. Sci. 2015, 16, 4865-4879. https://doi.org/10.3390/ijms16034865
Xu H, Lan D, Yang B, Wang Y. Biochemical Properties and Structure Analysis of a DAG-Like Lipase from Malassezia globosa. International Journal of Molecular Sciences. 2015; 16(3):4865-4879. https://doi.org/10.3390/ijms16034865
Chicago/Turabian StyleXu, Huan, Dongming Lan, Bo Yang, and Yonghua Wang. 2015. "Biochemical Properties and Structure Analysis of a DAG-Like Lipase from Malassezia globosa" International Journal of Molecular Sciences 16, no. 3: 4865-4879. https://doi.org/10.3390/ijms16034865
APA StyleXu, H., Lan, D., Yang, B., & Wang, Y. (2015). Biochemical Properties and Structure Analysis of a DAG-Like Lipase from Malassezia globosa. International Journal of Molecular Sciences, 16(3), 4865-4879. https://doi.org/10.3390/ijms16034865