Transcriptome Analysis of mRNA and miRNA in Somatic Embryos of Larix leptolepis Subjected to Hydrogen Treatment
Abstract
:1. Introduction
2. Results
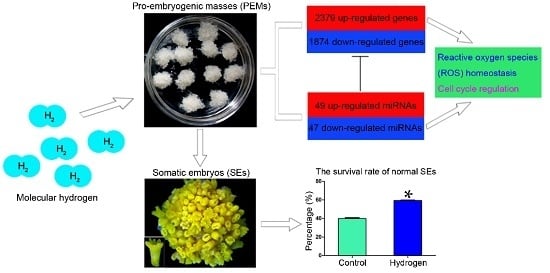
2.1. Hydrogen Improved the Survival Rates of Active PEMs and Normal SEs
2.2. H2 Reduced Reactive Oxygen Species (ROS) Levels and Enhanced Antioxidant Enzyme Activity
2.3. Changes in mRNA Expression in PEMs under Hydrogen Treatment
2.4. Validation of Differentially Expressed Genes (DEGs) by qPCR
2.5. Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) Enrichment Analysis
2.6. Effect of Hydrogen on miRNAs in Larch PEMs
2.7. mRNA-miRNA Interaction Network
3. Discussion
4. Experimental Section
4.1. Plant Materials and Growth Conditions
4.2. Hydrogen Treatment
4.3. Measurement of ROS Levels and Antioxidant Enzyme Activity
4.4. Microscopic Observation of PEMs and SEs
4.5. Library Preparation and Sequencing
4.6. De Novo Assembly and Statistical Analyses of mRNA-Seq Data
4.7. Real-Time Quantitative PCR (qPCR) Validation of DEGs
4.8. Statistical Analysis of miRNA-Seq Data
4.9. mRNA-miRNA Integration Analysis
4.10. Availability of Supporting Data
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
Abbreviations
| BBM | BABY BOOM |
| CAT | Catalase |
| DEG | Differentially Expressed Gene |
| FC | Fold Change |
| GO | Gene Ontology |
| KEGG | Kyoto Encyclopedia of Genes and Genomes |
| miRNA | MicroRNA |
| PCD | Programmed Cell Death |
| PEM | Pro-embryogenic Mass |
| POD | Peroxidase |
| qPCR | Real-time Quantitative PCR |
| ROS | Reactive Oxygen species |
| SE | Somatic Embryo |
| SOD | Superoxide Dismutase |
| TPM | Transcripts per Million |
| WUS | WUSCHEL |
References
- Ohsawa, I.; Ishikawa, M.; Takahashi, K.; Watanabe, M.; Nishimaki, K.; Yamagata, K.; Katsura, K.; Katayama, Y.; Asoh, S.; Ohta, S. Hydrogen acts as a therapeutic antioxidant by selectively reducing cytotoxic oxygen radicals. Nat. Med. 2007, 13, 688–694. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Sun, Y.P.; Li, Y.; Liu, W.W.; Xiang, H.G.; Fan, L.Y.; Sun, Q.; Xu, X.Y.; Cai, J.M.; Ruan, C.P.; et al. Hydrogen-rich saline ameliorates the severity of l-arginine-induced acute pancreatitis in rats. Biochem. Biophys. Res. Commun. 2010, 393, 308–313. [Google Scholar] [CrossRef] [PubMed]
- Fukuda, K.; Asoh, S.; Ishikawa, M.; Yamamoto, Y.; Ohsawa, I.; Ohta, S. Inhalation of hydrogen gas suppresses hepatic injury caused by ischemia/reperfusion through reducing oxidative stress. Biochem. Biophys. Res. Commun. 2007, 361, 670–674. [Google Scholar] [CrossRef] [PubMed]
- Hayashida, K.; Sano, M.; Ohsawa, I.; Shinmura, K.; Tamaki, K.; Kimura, K.; Endo, J.; Katayama, T.; Kawamura, A.; Kohsaka, S.; et al. Inhalation of hydrogen gas reduces infarct size in the rat model of myocardial ischemia-reperfusion injury. Biochem. Biophys. Res. Commun. 2008, 373, 30–35. [Google Scholar] [CrossRef] [PubMed]
- Itoh, T.; Fujita, Y.; Ito, M.; Masuda, A.; Ohno, K.; Ichihara, M.; Kojima, T.; Nozawa, Y. Molecular hydrogen suppresses FcεRI-mediated signal transduction and prevents degranulation of mast cells. Biochem. Biophys. Res. Commun. 2009, 389, 651–656. [Google Scholar] [CrossRef] [PubMed]
- Kajiyama, S.; Hasegawa, G.; Asano, M.; Hosoda, H.; Fukui, M.; Nakamura, N.; Kitawaki, J.; Imai, S.; Nakano, K.; Ohta, M.; et al. Supplementation of hydrogen-rich water improves lipid and glucose metabolism in patients with type 2 diabetes or impaired glucose tolerance. Nutr. Res. 2008, 28, 137–143. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Liu, K.; Sun, Q.; Liu, W.; Xu, W.; Denoble, P.; Tao, H.; Sun, X. Consumption of hydrogen water reduces paraquat-induced acute lung injury in rats. J. Biomed. Biotechnol. 2011, 2011, 305086. [Google Scholar] [CrossRef] [PubMed]
- Spulber, S.; Edoff, K.; Hong, L.; Morisawa, S.; Shirahata, S.; Ceccatelli, S. Molecular hydrogen reduces LPS-induced neuroinflammation and promotes recovery from sickness behaviour in mice. PLoS ONE 2012, 7, e42078. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.; Zhang, W.; Qi, F.; Cui, W.; Xie, Y.; Shen, W. Hydrogen-rich water regulates cucumber adventitious root development in a heme oxygenase-1/carbon monoxide-dependent manner. J. Plant Physiol. 2014, 171, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Xie, Y.; Mao, Y.; Zhang, W.; Lai, D.; Wang, Q.; Shen, W. Reactive oxygen species-dependent nitric oxide production contributes to hydrogen-promoted stomatal closure in Arabidopsis. Plant Physiol. 2014, 165, 759–773. [Google Scholar] [CrossRef] [PubMed]
- Xie, Y.; Mao, Y.; Lai, D.; Zhang, W.; Shen, W. H2 enhances Arabidopsis salt tolerance by manipulating ZAT10/12-mediated antioxidant defence and controlling sodium exclusion. PLoS ONE 2012, 7, e49800. [Google Scholar] [CrossRef] [PubMed]
- Xu, S.; Zhu, S.; Jiang, Y.; Wang, N.; Wang, R.; Shen, W.; Yang, J. Hydrogen-rich water alleviates salt stress in rice during seed germination. Plant Soil 2013, 370, 47–57. [Google Scholar] [CrossRef]
- Jin, Q.; Zhu, K.; Cui, W.; Xie, Y.; Han, B.; Shen, W. Hydrogen gas acts as a novel bioactive molecule in enhancing plant tolerance to paraquat-induced oxidative stress via the modulation of heme oxygenase-1 signalling system. Plant Cell Environ. 2013, 36, 956–969. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Zhao, X.; Wang, Z.; Shen, W.; Xu, X. Protective effects of hydrogen-rich water on the photosynthetic apparatus of maize seedlings (Zea mays L.) as a result of an increase in antioxidant enzyme activities under high light stress. Plant Growth Regul. 2015, 77, 43–56. [Google Scholar] [CrossRef]
- Gapper, C.; Dolan, L. Control of plant development by reactive oxygen species. Plant Physiol. 2006, 141, 341–345. [Google Scholar] [CrossRef] [PubMed]
- Miller, G.; Shulaev, V.; Mittler, R. Reactive oxygen signaling and abiotic stress. Physiol. Plant. 2008, 133, 481–489. [Google Scholar] [CrossRef] [PubMed]
- Gechev, T.S.; van Breusegem, F.; Stone, J.M.; Denev, I.; Laloi, C. Reactive oxygen species as signals that modulate plant stress responses and programmed cell death. Bioessays 2006, 28, 1091–1101. [Google Scholar] [CrossRef] [PubMed]
- Cui, W.; Fang, P.; Zhu, K.; Mao, Y.; Gao, C.; Xie, Y.; Wang, J.; Shen, W. Hydrogen-rich water confers plant tolerance to mercury toxicity in alfalfa seedlings. Ecotoxicol. Environ. Saf. 2014, 105, 103–111. [Google Scholar] [CrossRef] [PubMed]
- Cui, W.; Gao, C.; Fang, P.; Lin, G.; Shen, W. Alleviation of cadmium toxicity in Medicago sativa by hydrogen-rich water. J. Hazard. Mater. 2013, 260, 715–724. [Google Scholar] [CrossRef] [PubMed]
- Hu, H.; Li, P.; Wang, Y.; Gu, R. Hydrogen-rich water delays postharvest ripening and senescence of kiwifruit. Food Chem. 2014, 156, 100–109. [Google Scholar] [CrossRef] [PubMed]
- Wu, Q.; Su, N.; Cai, J.; Shen, Z.; Cui, J. Hydrogen-rich water enhances cadmium tolerance in Chinese cabbage by reducing cadmium uptake and increasing antioxidant capacities. J. Plant Physiol. 2015, 175, 174–182. [Google Scholar] [CrossRef] [PubMed]
- Von Arnold, S.; Sabala, I.; Bozhkov, P.; Dyachok, J.; Filonova, L. Developmental pathways of somatic embryogenesis. Plant Cell Tissue Organ Cult. 2002, 69, 233–249. [Google Scholar] [CrossRef]
- Filonova, L.H.; Bozhkov, P.V.; Brukhin, V.B.; Daniel, G.; Zhivotovsky, B.; von Arnold, S. Two waves of programmed cell death occur during formation and development of somatic embryos in the gymnosperm, norway spruce. J. Cell Sci. 2000, 113, 4399–4411. [Google Scholar] [PubMed]
- Li, S.G.; Li, W.F.; Han, S.Y.; Yang, W.H.; Qi, L.W. Stage-specific regulation of four HD-ZIP III transcription factors during polar pattern formation in Larix leptolepis somatic embryos. Gene 2013, 522, 177–183. [Google Scholar] [CrossRef] [PubMed]
- Li, W.F.; Zhang, S.G.; Han, S.Y.; Wu, T.; Zhang, J.H.; Qi, L.W. Regulation of LaMYB33 by miR159 during maintenance of embryogenic potential and somatic embryo maturation in larix kaempferi (lamb.) Carr. Plant Cell Tissue Organ Cult. 2013, 113, 131–136. [Google Scholar] [CrossRef]
- Zhang, J.; Wu, T.; Li, L.; Han, S.; Li, X.; Zhang, S.; Qi, L. Dynamic expression of small RNA populations in larch (Larix leptolepis). Planta 2013, 237, 89–101. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Zhang, S.; Han, S.; Li, X.; Tong, Z.; Qi, L. Deciphering small noncoding RNAs during the transition from dormant embryo to germinated embryo in larches (Larix leptolepis). PLoS ONE 2013, 8, e81452. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Zhang, S.; Han, S.; Wu, T.; Li, X.; Li, W.; Qi, L. Genome-wide identification of microRNAs in larch and stage-specific modulation of 11 conserved micrornas and their targets during somatic embryogenesis. Planta 2012, 236, 647–657. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Zhang, S.; Li, S.; Han, S.; Wu, T.; Li, X.; Qi, L. A genome-wide survey of microrna truncation and 3′ nucleotide addition events in larch (Larix leptolepis). Planta 2013, 237, 1047–1056. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Zhou, J.; Han, S.; Yang, W.; Li, W.; Wei, H.; Li, X.; Qi, L. Four abiotic stress-induced mirna families differentially regulated in the embryogenic and non-embryogenic callus tissues of Larix leptolepis. Biochem. Biophys. Res. Commun. 2010, 398, 355–360. [Google Scholar] [CrossRef] [PubMed]
- Bashandy, T.; Meyer, Y.; Reichheld, J.P. Redox regulation of auxin signaling and plant development in Arabidopsis. Plant Signal. Behav. 2011, 6, 117–119. [Google Scholar] [CrossRef] [PubMed]
- Zhou, T.; Yang, X.; Guo, K.; Deng, J.; Xu, J.; Gao, W.; Lindsey, K.; Zhang, X. ROS homeostasis regulates somatic embryogenesis via the regulation of auxin signaling in cotton. Mol. Cell. Proteom. 2016, 15, 2108–2124. [Google Scholar] [CrossRef] [PubMed]
- Boutilier, K.; Offringa, R.; Sharma, V.K.; Kieft, H.; Ouellet, T.; Zhang, L.; Hattori, J.; Liu, C.M.; van Lammeren, A.A.; Miki, B.L.; et al. Ectopic expression of BABY BOOM triggers a conversion from vegetative to embryonic growth. Plant Cell 2002, 14, 1737–1749. [Google Scholar] [CrossRef] [PubMed]
- El Ouakfaoui, S.; Schnell, J.; Abdeen, A.; Colville, A.; Labbe, H.; Han, S.; Baum, B.; Laberge, S.; Miki, B. Control of somatic embryogenesis and embryo development by AP2 transcription factors. Plant Mol. Biol. 2010, 74, 313–326. [Google Scholar] [CrossRef] [PubMed]
- Palovaara, J.; Hakman, I. Conifer WOX-related homeodomain transcription factors, developmental consideration and expression dynamic of WOX2 during picea abies somatic embryogenesis. Plant Mol. Biol. 2008, 66, 533–549. [Google Scholar] [CrossRef] [PubMed]
- Farage-Barhom, S.; Burd, S.; Sonego, L.; Perl-Treves, R.; Lers, A. Expression analysis of the BFN1 nuclease gene promoter during senescence, abscission, and programmed cell death-related processes. J. Exp. Bot. 2008, 59, 3247–3258. [Google Scholar] [CrossRef] [PubMed]
- Bollhöner, B.; Prestele, J.; Tuominen, H. Xylem cell death: Emerging understanding of regulation and function. J. Exp. Bot. 2012, 63, 1081–1094. [Google Scholar] [CrossRef] [PubMed]
- Hatsugai, N.; Yamada, K.; Goto-Yamada, S.; Hara-Nishimura, I. Vacuolar processing enzyme in plant programmed cell death. Front. Plant Sci. 2015, 6, 234. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tsiatsiani, L.; van Breusegem, F.; Gallois, P.; Zavialov, A.; Lam, E.; Bozhkov, P.V. Metacaspases. Cell Death Differ. 2011, 18, 1279–1288. [Google Scholar] [CrossRef] [PubMed]
- Maximova, S.N.; Florez, S.; Shen, X.; Niemenak, N.; Zhang, Y.; Curtis, W.; Guiltinan, M.J. Genome-wide analysis reveals divergent patterns of gene expression during zygotic and somatic embryomaturation of Theobroma cacao L., the chocolate tree. BMC Plant Biol. 2014, 14, 185. [Google Scholar] [CrossRef] [PubMed]
- Sun, G. MicroRNAs and their diverse functions in plants. Plant Mol. Biol. 2012, 80, 17–36. [Google Scholar] [CrossRef] [PubMed]
- Zhu, C.; Ding, Y.; Liu, H. MiR398 and plant stress responses. Physiol. Plant. 2011, 143, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Ma, C.; Burd, S.; Lers, A. miR408 is involved in abiotic stress responses in Arabidopsis. Plant J. 2015, 84, 169–187. [Google Scholar] [CrossRef] [PubMed]
- Jia, N.; Lv, T.T.; Li, M.X.; Wei, S.S.; Li, Y.Y.; Zhao, C.L.; Li, B. The J-protein AtDjB1 is required for mitochondrial complex i activity and regulates growth and development through ROS-mediated auxin signalling. J. Exp. Bot. 2016, 67, 3481–3496. [Google Scholar] [CrossRef] [PubMed]
- Seo, T.; Kurokawa, R.; Sato, B. A convenient method for determining the concentration of hydrogen in water: Use of methylene blue with colloidal platinum. Med. Gas Res. 2012, 2, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Kristiansen, K.A.; Jensen, P.E.; Moller, I.M.; Schulz, A. Monitoring reactive oxygen species formation and localisation in living cells by use of the fluorescent probe CM-H2DCFDA and confocal laser microscopy. Physiol. Plant. 2009, 136, 369–383. [Google Scholar] [CrossRef] [PubMed]
- Beauchamp, C.; Fridovich, I. Superoxide dismutase: Improved assays and an assay applicable to acrylamide gels. Anal. Biochem. 1971, 44, 276–287. [Google Scholar] [CrossRef]
- Aebi, H. Catalase in vitro. Methods Enzymol. 1984, 105, 121–126. [Google Scholar] [PubMed]
- Rao, M.V.; Paliyath, G.; Ormrod, D.P. Ultraviolet-B- and ozone-induced biochemical changes in antioxidant enzymes of Arabidopsis thaliana. Plant Physiol. 1996, 110, 125–136. [Google Scholar] [CrossRef] [PubMed]
- Gupta, P.K.; Dandekar, A.M.; Durzan, D.J. Somatic proembryo formation and transient expression of a luciferase gene in Douglas fir and loblolly pine protoplasts. Plant Sci. 1988, 58, 85–92. [Google Scholar] [CrossRef]
- Haas, B.J.; Papanicolaou, A.; Yassour, M.; Grabherr, M.; Blood, P.D.; Bowden, J.; Couger, M.B.; Eccles, D.; Li, B.; Lieber, M.; et al. De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat. Protoc. 2013, 8, 1494–1512. [Google Scholar] [CrossRef] [PubMed]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Dewey, C.N. RSEM: Accurate transcript quantification from RNA-seq data with or without a reference genome. BMC Bioinform. 2011, 12, 323. [Google Scholar] [CrossRef] [PubMed]
- Anders, S.; Huber, W. Differential expression analysis for sequence count data. Genome Biol. 2010, 11, R106. [Google Scholar] [CrossRef] [PubMed]
- Young, M.D.; Wakefield, M.J.; Smyth, G.K.; Oshlack, A. Gene ontology analysis for RNA-seq: Accounting for selection bias. Genome Biol. 2010, 11, R14. [Google Scholar] [CrossRef] [PubMed]
- Xie, C.; Mao, X.; Huang, J.; Ding, Y.; Wu, J.; Dong, S.; Kong, L.; Gao, G.; Li, C.Y.; Wei, L. KOBAS 2.0: A web server for annotation and identification of enriched pathways and diseases. Nucleic Acids Res. 2011, 39, W316–W322. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCt method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Nystedt, B.; Street, N.R.; Wetterbom, A.; Zuccolo, A.; Lin, Y.C.; Scofield, D.G.; Vezzi, F.; Delhomme, N.; Giacomello, S.; Alexeyenko, A.; et al. The norway spruce genome sequence and conifer genome evolution. Nature 2013, 497, 579–584. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Friedlander, M.R.; Mackowiak, S.D.; Li, N.; Chen, W.; Rajewsky, N. miRDeep2 accurately identifies known and hundreds of novel microrna genes in seven animal clades. Nucleic Acids Res. 2012, 40, 37–52. [Google Scholar] [CrossRef] [PubMed]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. edgeR: A bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.J.; Ma, Y.K.; Chen, T.; Wang, M.; Wang, X.J. PsRobot: A web-based plant small RNA meta-analysis toolbox. Nucleic Acids Res. 2012, 40, W22–W28. [Google Scholar] [CrossRef] [PubMed]
- Dessau, R.B.; Pipper, C.B. [“R”--project for statistical computing]. Ugeskr. Laeger 2008, 170, 328–330. [Google Scholar] [PubMed]






© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, Y.; Han, S.; Ding, X.; Li, X.; Zhang, L.; Li, W.; Xu, H.; Li, Z.; Qi, L. Transcriptome Analysis of mRNA and miRNA in Somatic Embryos of Larix leptolepis Subjected to Hydrogen Treatment. Int. J. Mol. Sci. 2016, 17, 1951. https://doi.org/10.3390/ijms17111951
Liu Y, Han S, Ding X, Li X, Zhang L, Li W, Xu H, Li Z, Qi L. Transcriptome Analysis of mRNA and miRNA in Somatic Embryos of Larix leptolepis Subjected to Hydrogen Treatment. International Journal of Molecular Sciences. 2016; 17(11):1951. https://doi.org/10.3390/ijms17111951
Chicago/Turabian StyleLiu, Yali, Suying Han, Xiangming Ding, Xinmin Li, Lifeng Zhang, Wanfeng Li, Haiyan Xu, Zhexin Li, and Liwang Qi. 2016. "Transcriptome Analysis of mRNA and miRNA in Somatic Embryos of Larix leptolepis Subjected to Hydrogen Treatment" International Journal of Molecular Sciences 17, no. 11: 1951. https://doi.org/10.3390/ijms17111951
APA StyleLiu, Y., Han, S., Ding, X., Li, X., Zhang, L., Li, W., Xu, H., Li, Z., & Qi, L. (2016). Transcriptome Analysis of mRNA and miRNA in Somatic Embryos of Larix leptolepis Subjected to Hydrogen Treatment. International Journal of Molecular Sciences, 17(11), 1951. https://doi.org/10.3390/ijms17111951