MicroRNAs, DNA Damage Response, and Cancer Treatment
Abstract
:1. Introduction
1.1. DNA Damage Response (DDR) and Cancer
1.2. Biogenesis and Function of MicroRNAs (miRNAs)
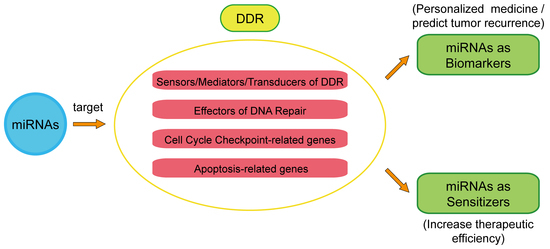
2. miRNAs Mediate DDR Regulation
2.1. Sensors/Mediators/Transducers of DDR
2.2. Effectors of DNA Repair
2.3. Effectors of Apoptosis and Cell Cycle Checkpoint
3. miRNA’s Therapeutic Function in Cancer Based on Its Regulation of the DDR
3.1. DDR-Related miRNAs as Biomarkers in Clinical Cancer Therapy
3.2. DDR-Related miRNAs as Sensitizers to Radiotherapy or Chemotherapy
4. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Lindahl, T.; Barnes, D.E. Repair of endogenous DNA damage. Cold Spring Harb. Symp. Quant. Biol. 2000, 65, 127–133. [Google Scholar] [CrossRef] [PubMed]
- Hoeijmakers, J.H. DNA damage, aging, and cancer. N. Eng. J. Med. 2009, 361, 1475–1485. [Google Scholar] [CrossRef] [PubMed]
- Khanna, K.K.; Jackson, S.P. DNA double-strand breaks: Signaling, repair and the cancer connection. Nat. Genet. 2001, 27, 247–254. [Google Scholar] [CrossRef] [PubMed]
- Tian, H.; Gao, Z.; Li, H.; Zhang, B.; Wang, G.; Zhang, Q.; Pei, D.; Zheng, J. DNA damage response—A double-edged sword in cancer prevention and cancer therapy. Cancer Lett. 2015, 358, 8–16. [Google Scholar] [CrossRef] [PubMed]
- Bartel, D.P. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell 2004, 116, 281–297. [Google Scholar] [CrossRef]
- Lee, R.C.; Feinbaum, R.L.; Ambros, V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 1993, 75, 843–854. [Google Scholar] [CrossRef]
- Carthew, R.W.; Sontheimer, E.J. Origins and mechanisms of miRNAs and siRNAs. Cell 2009, 136, 642–655. [Google Scholar] [CrossRef] [PubMed]
- Bushati, N.; Cohen, S.M. MicroRNA functions. Annu. Rev. Cell Dev. Biol. 2007, 23, 175–205. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.; Ahn, C.; Han, J.; Choi, H.; Kim, J.; Yim, J.; Lee, J.; Provost, P.; Radmark, O.; Kim, S.; et al. The nuclear RNase III Drosha initiates microRNA processing. Nature 2003, 425, 415–419. [Google Scholar] [CrossRef] [PubMed]
- Yi, R.; Qin, Y.; Macara, I.G.; Cullen, B.R. Exportin-5 mediates the nuclear export of pre-microRNAs and short hairpin RNAs. Genes Dev. 2003, 17, 3011–3016. [Google Scholar] [CrossRef] [PubMed]
- Cai, X.; Hagedorn, C.H.; Cullen, B.R. Human microRNAs are processed from capped, polyadenylated transcripts that can also function as mRNAs. RNA 2004, 10, 1957–1966. [Google Scholar] [CrossRef] [PubMed]
- Macfarlane, L.A.; Murphy, P.R. MicroRNA: Biogenesis, function and role in cancer. Curr. Genom. 2010, 11, 537–561. [Google Scholar] [CrossRef] [PubMed]
- Han, C.; Wan, G.; Langley, R.R.; Zhang, X.; Lu, X. Crosstalk between the DNA damage response pathway and microRNAs. Cell. Mol. Life Sci. 2012, 69, 2895–2906. [Google Scholar] [CrossRef] [PubMed]
- Wan, G.; Mathur, R.; Hu, X.; Zhang, X.; Lu, X. miRNA response to DNA damage. Trends Biochem. Sci. 2011, 36, 478–484. [Google Scholar] [CrossRef] [PubMed]
- Hu, H.; Gatti, R.A. MicroRNAs: New players in the DNA damage response. J. Mol. Cell Biol. 2011, 3, 151–158. [Google Scholar] [CrossRef] [PubMed]
- Harper, J.W.; Elledge, S.J. The DNA damage response: Ten years after. Mol. Cell 2007, 28, 739–745. [Google Scholar] [CrossRef] [PubMed]
- Goldstein, M.; Kastan, M.B. The DNA damage response: Implications for tumor responses to radiation and chemotherapy. Annu. Rev. Med. 2015, 66, 129–143. [Google Scholar] [CrossRef] [PubMed]
- Lal, A.; Pan, Y.; Navarro, F.; Dykxhoorn, D.M.; Moreau, L.; Meire, E.; Bentwich, Z.; Lieberman, J.; Chowdhury, D. miR-24-mediated downregulation of H2AX suppresses DNA repair in terminally differentiated blood cells. Nat. Struct. Mol. Biol. 2009, 16, 492–498. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.; Luo, J.; Liu, Z.; Zhou, R.; Luo, H. MicroRNA-138 regulates DNA damage response in small cell lung cancer cells by directly targeting H2AX. Cancer Investig. 2015, 33, 126–136. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Huang, J.W.; Li, M.; Cavenee, W.K.; Mitchell, P.S.; Zhou, X.; Tewari, M.; Furnari, F.B.; Taniguchi, T. MicroRNA-138 modulates DNA damage response by repressing histone H2AX expression. Mol. Cancer Res. 2011, 9, 1100–1111. [Google Scholar] [CrossRef] [PubMed]
- Song, L.; Lin, C.; Wu, Z.; Gong, H.; Zeng, Y.; Wu, J.; Li, M.; Li, J. miR-18a impairs DNA damage response through downregulation of ataxia telangiectasia mutated (ATM) kinase. PLoS ONE 2011, 6, e25454. [Google Scholar] [CrossRef] [PubMed]
- Mansour, W.Y.; Bogdanova, N.V.; Kasten-Pisula, U.; Rieckmann, T.; Kocher, S.; Borgmann, K.; Baumann, M.; Krause, M.; Petersen, C.; Hu, H.; et al. Aberrant overexpression of miR-421 downregulates ATM and leads to a pronounced DSB repair defect and clinical hypersensitivity in SKX squamous cell carcinoma. Radiother. Oncol. 2013, 106, 147–154. [Google Scholar] [CrossRef] [PubMed]
- Yan, D.; Ng, W.L.; Zhang, X.; Wang, P.; Zhang, Z.; Mo, Y.Y.; Mao, H.; Hao, C.; Olson, J.J.; Curran, W.J.; et al. Targeting DNA-pkcs and ATM with miR-101 sensitizes tumors to radiation. PLoS ONE 2010, 5, e11397. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Wan, G.; Spizzo, R.; Ivan, C.; Mathur, R.; Hu, X.; Ye, X.; Lu, J.; Fan, F.; Xia, L.; et al. miR-203 induces oxaliplatin resistance in colorectal cancer cells by negatively regulating ATM kinase. Mol. Oncol. 2014, 8, 83–92. [Google Scholar] [CrossRef] [PubMed]
- Guo, X.; Yang, C.; Qian, X.; Lei, T.; Li, Y.; Shen, H.; Fu, L.; Xu, B. Estrogen receptor α regulates ATM expression through miRNAs in breast cancer. Clin. Cancer Res. 2013, 19, 4994–5002. [Google Scholar] [CrossRef] [PubMed]
- Di Francesco, A.; de Pitta, C.; Moret, F.; Barbieri, V.; Celotti, L.; Mognato, M. The DNA-damage response to γ-radiation is affected by miR-27a in A549 cells. Int. J. Mol. Sci. 2013, 14, 17881–17896. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; He, J.; Su, F.; Ding, N.; Hu, W.; Yao, B.; Wang, W.; Zhou, G. Repression of atr pathway by miR-185 enhances radiation-induced apoptosis and proliferation inhibition. Cell Death Dis. 2013, 4, e699. [Google Scholar] [CrossRef] [PubMed]
- Sun, C.; Li, N.; Yang, Z.; Zhou, B.; He, Y.; Weng, D.; Fang, Y.; Wu, P.; Chen, P.; Yang, X.; et al. miR-9 regulation of BRCA1 and ovarian cancer sensitivity to cisplatin and PARP inhibition. J. Natl. Cancer Inst. 2013, 105, 1750–1758. [Google Scholar] [CrossRef] [PubMed]
- Lal, A.; Navarro, F.; Maher, C.A.; Maliszewski, L.E.; Yan, N.; O’Day, E.; Chowdhury, D.; Dykxhoorn, D.M.; Tsai, P.; Hofmann, O.; et al. miR-24 inhibits cell proliferation by targeting E2F2, MYC, and other cell-cycle genes via binding to “seedless” 3’UTR microRNA recognition elements. Mol. Cell 2009, 35, 610–625. [Google Scholar] [CrossRef] [PubMed]
- Moskwa, P.; Buffa, F.M.; Pan, Y.; Panchakshari, R.; Gottipati, P.; Muschel, R.J.; Beech, J.; Kulshrestha, R.; Abdelmohsen, K.; Weinstock, D.M.; et al. miR-182-mediated downregulation of BRCA1 impacts DNA repair and sensitivity to PARP inhibitors. Mol. Cell 2011, 41, 210–220. [Google Scholar] [CrossRef] [PubMed]
- Johnson, C.D.; Esquela-Kerscher, A.; Stefani, G.; Byrom, M.; Kelnar, K.; Ovcharenko, D.; Wilson, M.; Wang, X.; Shelton, J.; Shingara, J.; et al. The let-7 microRNA represses cell proliferation pathways in human cells. Cancer Res. 2007, 67, 7713–7722. [Google Scholar] [CrossRef] [PubMed]
- Liu, R.L.; Dong, Y.; Deng, Y.Z.; Wang, W.J.; Li, W.D. Tumor suppressor miR-145 reverses drug resistance by directly targeting DNA damage-related gene RAD18 in colorectal cancer. Tumour Biol. 2015, 36, 5011–5019. [Google Scholar] [CrossRef] [PubMed]
- Cortez, M.A.; Valdecanas, D.; Niknam, S.; Peltier, H.J.; Diao, L.; Giri, U.; Komaki, R.; Calin, G.A.; Gomez, D.R.; Chang, J.Y.; et al. In vivo delivery of miR-34a sensitizes lung tumors to radiation through RAD51 regulation. Mol. Ther.—Nucleic Acids 2015, 4, e270. [Google Scholar] [CrossRef] [PubMed]
- Liu, G.; Yang, D.; Rupaimoole, R.; Pecot, C.V.; Sun, Y.; Mangala, L.S.; Li, X.; Ji, P.; Cogdell, D.; Hu, L.; et al. Augmentation of response to chemotherapy by microRNA-506 through regulation of RAD51 in serous ovarian cancers. J. Natl. Cancer Inst. 2015. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.W.; Wang, Y.; Dhillon, K.K.; Calses, P.; Villegas, E.; Mitchell, P.S.; Tewari, M.; Kemp, C.J.; Taniguchi, T. Systematic screen identifies miRNAs that target RAD51 and RAD51D to enhance chemosensitivity. Mol. Cancer Res. 2013, 11, 1564–1573. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Huang, J.W.; Calses, P.; Kemp, C.J.; Taniguchi, T. miR-96 downregulates REV1 and RAD51 to promote cellular sensitivity to cisplatin and PARP inhibition. Cancer Res. 2012, 72, 4037–4046. [Google Scholar] [CrossRef] [PubMed]
- Gasparini, P.; Lovat, F.; Fassan, M.; Casadei, L.; Cascione, L.; Jacob, N.K.; Carasi, S.; Palmieri, D.; Costinean, S.; Shapiro, C.L.; et al. Protective role of miR-155 in breast cancer through RAD51 targeting impairs homologous recombination after irradiation. Proc. Natl. Acad. Sci. USA 2014, 111, 4536–4541. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Fu, W.; Liao, H.; Dai, L.; Jiang, Z.; Pan, Y.; Huang, H.; Mo, Y.; Li, S.; Yang, G.; et al. The regulatory and predictive functions of miR-17 and miR-92 families on cisplatin resistance of non-small cell lung cancer. BMC Cancer 2015. [Google Scholar] [CrossRef] [PubMed]
- Crosby, M.E.; Kulshreshtha, R.; Ivan, M.; Glazer, P.M. MicroRNA regulation of DNA repair gene expression in hypoxic stress. Cancer Res. 2009, 69, 1221–1229. [Google Scholar] [CrossRef] [PubMed]
- Zhang, P.; Wang, L.; Rodriguez-Aguayo, C.; Yuan, Y.; Debeb, B.G.; Chen, D.; Sun, Y.; You, M.J.; Liu, Y.; Dean, D.C.; et al. miR-205 acts as a tumour radiosensitizer by targeting ZEB1 and UBC13. Nat. Commun. 2014. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Hu, G.; Li, P.; Tang, S.; Zhang, J.; Jia, G. miR-3940–5p enhances homologous recombination after DSB in Cr(VI) exposed 16HBE cell. Toxicology 2016, 344, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Hatano, K.; Kumar, B.; Zhang, Y.; Coulter, J.B.; Hedayati, M.; Mears, B.; Ni, X.; Kudrolli, T.A.; Chowdhury, W.H.; Rodriguez, R.; et al. A functional screen identifies miRNAs that inhibit DNA repair and sensitize prostate cancer cells to ionizing radiation. Nucleic Acids Res. 2015, 43, 4075–4086. [Google Scholar] [CrossRef] [PubMed]
- Pouliot, L.M.; Chen, Y.C.; Bai, J.; Guha, R.; Martin, S.E.; Gottesman, M.M.; Hall, M.D. Cisplatin sensitivity mediated by WEE1 and CHK1 is mediated by miR-155 and the miR-15 family. Cancer Res. 2012, 72, 5945–5955. [Google Scholar] [CrossRef] [PubMed]
- Soriano, A.; Paris-Coderch, L.; Jubierre, L.; Martinez, A.; Zhou, X.; Piskareva, O.; Bray, I.; Vidal, I.; Almazan-Moga, A.; Molist, C.; et al. MicroRNA-497 impairs the growth of chemoresistant neuroblastoma cells by targeting cell cycle, survival and vascular permeability genes. Oncotarget 2016, 7, 9271–9287. [Google Scholar] [PubMed]
- Galluzzi, L.; Morselli, E.; Vitale, I.; Kepp, O.; Senovilla, L.; Criollo, A.; Servant, N.; Paccard, C.; Hupe, P.; Robert, T.; et al. miR-181A and miR-630 regulate cisplatin-induced cancer cell death. Cancer Res. 2010, 70, 1793–1803. [Google Scholar] [CrossRef] [PubMed]
- Hu, W.; Chan, C.S.; Wu, R.; Zhang, C.; Sun, Y.; Song, J.S.; Tang, L.H.; Levine, A.J.; Feng, Z. Negative regulation of tumor suppressor p53 by microRNA miR-504. Mol. Cell 2010, 38, 689–699. [Google Scholar] [CrossRef] [PubMed]
- Zhou, M.; Liu, Z.; Zhao, Y.; Ding, Y.; Liu, H.; Xi, Y.; Xiong, W.; Li, G.; Lu, J.; Fodstad, O.; et al. MicroRNA-125b confers the resistance of breast cancer cells to paclitaxel through suppression of pro-apoptotic Bcl-2 antagonist killer 1 (BAK1) expression. J. Biol. Chem. 2010, 285, 21496–21507. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Xing, R.; Zhang, X.; Dong, W.; Zhang, J.; Yan, Z.; Li, W.; Cui, J.; Lu, Y. miR-375 targets the p53 gene to regulate cellular response to ionizing radiation and etoposide in gastric cancer cells. DNA Repair 2013, 12, 741–750. [Google Scholar] [CrossRef] [PubMed]
- Ivanovska, I.; Ball, A.S.; Diaz, R.L.; Magnus, J.F.; Kibukawa, M.; Schelter, J.M.; Kobayashi, S.V.; Lim, L.; Burchard, J.; Jackson, A.L.; et al. MicroRNAs in the miR-106B family regulate p21/CDKN1A and promote cell cycle progression. Mol. Cell. Biol. 2008, 28, 2167–2174. [Google Scholar] [CrossRef] [PubMed]
- Nakamachi, Y.; Kawano, S.; Takenokuchi, M.; Nishimura, K.; Sakai, Y.; Chin, T.; Saura, R.; Kurosaka, M.; Kumagai, S. MicroRNA-124a is a key regulator of proliferation and monocyte chemoattractant protein 1 secretion in fibroblast-like synoviocytes from patients with rheumatoid arthritis. Arthritis Rheum. 2009, 60, 1294–1304. [Google Scholar] [CrossRef] [PubMed]
- Afanasyeva, E.A.; Mestdagh, P.; Kumps, C.; Vandesompele, J.; Ehemann, V.; Theissen, J.; Fischer, M.; Zapatka, M.; Brors, B.; Savelyeva, L.; et al. MicroRNA miR-885–5p targets CDK2 and MCM5, activates p53 and inhibits proliferation and survival. Cell Death Diff. 2011, 18, 974–984. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Lv, Q.; He, J.; Zhang, H.; Mei, X.; Cui, K.; Huang, N.; Xie, W.; Xu, N.; Zhang, Y. MicroRNA-188 suppresses G1/S transition by targeting multiple cyclin/CDK complexes. Cell Commun. Signal. 2014, 12, 66. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Huang, W.; Ran, Y.; Xiong, Y.; Zhong, Z.; Fan, X.; Wang, Z.; Ye, Q. miR-582–5p inhibits proliferation of hepatocellular carcinoma by targeting CDK1 and AKT3. Tumour Biol. 2015, 36, 8309–8316. [Google Scholar] [CrossRef] [PubMed]
- Lin, J.; Liu, C.; Gao, F.; Mitchel, R.E.; Zhao, L.; Yang, Y.; Lei, J.; Cai, J. miR-200c enhances radiosensitivity of human breast cancer cells. J. Cell. Biochem. 2013, 114, 606–615. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Kong, Y.; Xu, X.; Xing, H.; Zhang, Y.; Han, F.; Li, W.; Yang, Q.; Zeng, J.; Jia, J.; et al. F-box protein FBXO31 is down-regulated in gastric cancer and negatively regulated by miR-17 and miR-20a. Oncotarget 2014, 5, 6178–6190. [Google Scholar] [CrossRef] [PubMed]
- Pothof, J.; Verkaik, N.S.; van, I.W.; Wiemer, E.A.; Ta, V.T.; van der Horst, G.T.; Jaspers, N.G.; van Gent, D.C.; Hoeijmakers, J.H.; Persengiev, S.P. MicroRNA-mediated gene silencing modulates the UV-induced DNA-damage response. EMBO J. 2009, 28, 2090–2099. [Google Scholar] [CrossRef] [PubMed]
- De Oliveira, P.E.; Zhang, L.; Wang, Z.; Lazo, J.S. Hypoxia-mediated regulation of CDC25A phosphatase by p21 and miR-21. Cell Cycle 2009, 8, 3157–3164. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Feng, M.; Jiang, X.; Wu, Z.; Li, Z.; Aau, M.; Yu, Q. miR-449a and miR-449b are direct transcriptional targets of E2F1 and negatively regulate PRB-E2F1 activity through a feedback loop by targeting CDK6 and CDC25A. Genes Dev. 2009, 23, 2388–2393. [Google Scholar] [CrossRef] [PubMed]
- He, M.; Lin, Y.; Tang, Y.; Liu, Y.; Zhou, W.; Li, C.; Sun, G.; Guo, M. miR-638 suppresses DNA damage repair by targeting SMC1A expression in terminally differentiated cells. Aging 2016, 8, 1442–1456. [Google Scholar] [CrossRef] [PubMed]
- Alla, V.; Kowtharapu, B.S.; Engelmann, D.; Emmrich, S.; Schmitz, U.; Steder, M.; Putzer, B.M. E2F1 confers anticancer drug resistance by targeting ABC transporter family members and Bcl-2 via the p73/DNP73-miR-205 circuitry. Cell Cycle 2012, 11, 3067–3078. [Google Scholar] [CrossRef] [PubMed]
- Tsang, W.P.; Kwok, T.T. Epigallocatechin gallate up-regulation of miR-16 and induction of apoptosis in human cancer cells. J. Nutr. Biochem. 2010, 21, 140–146. [Google Scholar] [CrossRef] [PubMed]
- Mei, Z.; Su, T.; Ye, J.; Yang, C.; Zhang, S.; Xie, C. The miR-15 family enhances the radiosensitivity of breast cancer cells by targeting G2 checkpoints. Radiat. Res. 2015, 183, 196–207. [Google Scholar] [CrossRef] [PubMed]
- Kwon, J.E.; Kim, B.Y.; Kwak, S.Y.; Bae, I.H.; Han, Y.H. Ionizing radiation-inducible microRNA miR-193a-3p induces apoptosis by directly targeting MCL-1. Apoptosis 2013, 18, 896–909. [Google Scholar] [CrossRef] [PubMed]
- Zang, Y.S.; Zhong, Y.F.; Fang, Z.; Li, B.; An, J. miR-155 inhibits the sensitivity of lung cancer cells to cisplatin via negative regulation of APAF-1 expression. Cancer Gene Ther. 2012, 19, 773–778. [Google Scholar] [CrossRef] [PubMed]
- Niu, J.; Shi, Y.; Tan, G.; Yang, C.H.; Fan, M.; Pfeffer, L.M.; Wu, Z.H. DNA damage induces NF-κB-dependent microRNA-21 up-regulation and promotes breast cancer cell invasion. J. Biol. Chem. 2012, 287, 21783–21795. [Google Scholar] [CrossRef] [PubMed]
- Lieber, M.R.; Ma, Y.; Pannicke, U.; Schwarz, K. Mechanism and regulation of human non-homologous DNA end-joining. Nat. Rev. 2003, 4, 712–720. [Google Scholar] [CrossRef] [PubMed]
- Zhu, F.; Liu, J.L.; Li, J.P.; Xiao, F.; Zhang, Z.X.; Zhang, L. MicroRNA-124 (miR-124) regulates Ku70 expression and is correlated with neuronal death induced by ischemia/reperfusion. J. Mol. Neurosci. 2014, 52, 148–155. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.Y.; Fu, S.L.; Xu, S.Q.; Zhou, X.; Liu, X.S.; Xu, Y.J.; Zhao, J.P.; Wei, S. By downregulating Ku80, HSA-miR-526b suppresses non-small cell lung cancer. Oncotarget 2015, 6, 1462–1477. [Google Scholar] [CrossRef] [PubMed]
- Ciccia, A.; Elledge, S.J. The DNA damage response: Making it safe to play with knives. Mol. Cell 2010, 40, 179–204. [Google Scholar] [CrossRef] [PubMed]
- Neijenhuis, S.; Bajrami, I.; Miller, R.; Lord, C.J.; Ashworth, A. Identification of miRNA modulators to PARP inhibitor response. DNA Repair 2013, 12, 394–402. [Google Scholar] [CrossRef] [PubMed]
- Mueller, A.C.; Sun, D.; Dutta, A. The miR-99 family regulates the DNA damage response through its target SNF2H. Oncogene 2013, 32, 1164–1172. [Google Scholar] [CrossRef] [PubMed]
- Zhao, L.; Bode, A.M.; Cao, Y.; Dong, Z. Regulatory mechanisms and clinical perspectives of miRNA in tumor radiosensitivity. Carcinogenesis 2012, 33, 2220–2227. [Google Scholar] [CrossRef] [PubMed]
- Kumar, M.; Lu, Z.; Takwi, A.A.; Chen, W.; Callander, N.S.; Ramos, K.S.; Young, K.H.; Li, Y. Negative regulation of the tumor suppressor p53 gene by microRNAs. Oncogene 2011, 30, 843–853. [Google Scholar] [CrossRef] [PubMed]
- Hong, C.F.; Lin, S.Y.; Chou, Y.T.; Wu, C.W. MicroRNA-7 compromises p53 protein-dependent apoptosis by controlling the expression of the chromatin remodeling factor SMARCD1. J. Biol. Chem. 2016, 291, 1877–1889. [Google Scholar] [CrossRef] [PubMed]
- Rosenfeld, N.; Aharonov, R.; Meiri, E.; Rosenwald, S.; Spector, Y.; Zepeniuk, M.; Benjamin, H.; Shabes, N.; Tabak, S.; Levy, A.; et al. MicroRNAs accurately identify cancer tissue origin. Nat. Biotechnol. 2008, 26, 462–469. [Google Scholar] [CrossRef] [PubMed]
- Lu, J.; Getz, G.; Miska, E.A.; Alvarez-Saavedra, E.; Lamb, J.; Peck, D.; Sweet-Cordero, A.; Ebert, B.L.; Mak, R.H.; Ferrando, A.A.; et al. MicroRNA expression profiles classify human cancers. Nature 2005, 435, 834–838. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Wu, S.; Shi, Y.; Miao, Y.; Luo, X.; Ji, M.; Yao, K.; He, J. c-MYB regulates cell growth and DNA damage repair through modulating miR-143. FEBS Lett. 2015, 589, 555–564. [Google Scholar] [CrossRef] [PubMed]
- Besse, A.; Sana, J.; Lakomy, R.; Kren, L.; Fadrus, P.; Smrcka, M.; Hermanova, M.; Jancalek, R.; Reguli, S.; Lipina, R.; et al. miR-338–5p sensitizes glioblastoma cells to radiation through regulation of genes involved in DNA damage response. Tumour Biol. 2016, 37, 7719–7727. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Pan, X.; Yang, Q.; Wen, L.; Jiang, Y.; Zhao, Y.; Li, G. MicroRNA-18a enhances the radiosensitivity of cervical cancer cells by promoting radiation-induced apoptosis. Oncol. Rep. 2015, 33, 2853–2862. [Google Scholar] [CrossRef] [PubMed]
- Lynam-Lennon, N.; Reynolds, J.V.; Marignol, L.; Sheils, O.M.; Pidgeon, G.P.; Maher, S.G. MicroRNA-31 modulates tumour sensitivity to radiation in oesophageal adenocarcinoma. J. Mol. Med. 2012, 90, 1449–1458. [Google Scholar] [CrossRef] [PubMed]
- Ebi, H.; Sato, T.; Sugito, N.; Hosono, Y.; Yatabe, Y.; Matsuyama, Y.; Yamaguchi, T.; Osada, H.; Suzuki, M.; Takahashi, T. Counterbalance between RB inactivation and miR-17–92 overexpression in reactive oxygen species and DNA damage induction in lung cancers. Oncogene 2009, 28, 3371–3379. [Google Scholar] [CrossRef] [PubMed]
- Ohyashiki, K.; Umezu, T.; Yoshizawa, S.; Ito, Y.; Ohyashiki, M.; Kawashima, H.; Tanaka, M.; Kuroda, M.; Ohyashiki, J.H. Clinical impact of down-regulated plasma miR-92a levels in non-hodgkin’s lymphoma. PLoS ONE 2011, 6, e16408. [Google Scholar] [CrossRef] [PubMed]
- Ferrajoli, A.; Shanafelt, T.D.; Ivan, C.; Shimizu, M.; Rabe, K.G.; Nouraee, N.; Ikuo, M.; Ghosh, A.K.; Lerner, S.; Rassenti, L.Z.; et al. Prognostic value of miR-155 in individuals with monoclonal B-cell lymphocytosis and patients with B chronic lymphocytic leukemia. Blood 2013, 122, 1891–1899. [Google Scholar] [CrossRef] [PubMed]
- Rebucci, M.; Michiels, C. Molecular aspects of cancer cell resistance to chemotherapy. Biochem. Pharm. 2013, 85, 1219–1226. [Google Scholar] [CrossRef] [PubMed]
- He, J.; Yu, J.J.; Xu, Q.; Wang, L.; Zheng, J.Z.; Liu, L.Z.; Jiang, B.H. Downregulation of ATG14 by EGR1-miR152 sensitizes ovarian cancer cells to cisplatin-induced apoptosis by inhibiting cyto-protective autophagy. Autophagy 2015, 11, 373–384. [Google Scholar] [CrossRef] [PubMed]
- Kastl, L.; Brown, I.; Schofield, A.C. miRNA-34a is associated with docetaxel resistance in human breast cancer cells. Breast Cancer Res. Treat. 2012, 131, 445–454. [Google Scholar] [CrossRef] [PubMed]
- Xu, R.X.; Liu, R.Y.; Wu, C.M.; Zhao, Y.S.; Li, Y.; Yao, Y.Q.; Xu, Y.H. DNA damage-induced NF-κB activation in human glioblastoma cells promotes miR-181b expression and cell proliferation. Cell. Physiol. Biochem. 2015, 35, 913–925. [Google Scholar] [CrossRef] [PubMed]
- Ren, J.; Chu, Y.; Ma, H.; Zhang, Y.; Zhang, X.; Zhao, D.; Li, Z.; Wang, J.; Gao, Y.E.; Xiao, L.; et al. Epigenetic interventions increase the radiation sensitivity of cancer cells. Curr. Pharm. Des. 2014, 20, 1857–1865. [Google Scholar] [CrossRef] [PubMed]
- Zhu, W.; Zhu, D.; Lu, S.; Wang, T.; Wang, J.; Jiang, B.; Shu, Y.; Liu, P. miR-497 modulates multidrug resistance of human cancer cell lines by targeting Bcl2. Med. Oncol. 2012, 29, 384–391. [Google Scholar] [CrossRef] [PubMed]
- Fu, W.F.; Chen, W.B.; Dai, L.; Yang, G.P.; Jiang, Z.Y.; Pan, L.; Zhao, J.; Chen, G. Inhibition of miR-141 reverses cisplatin resistance in non-small cell lung cancer cells via upregulation of programmed cell death protein 4. Eur. Rev. Med. Pharm. Sci. 2016, 20, 2565–2572. [Google Scholar]
- Samuel, P.; Pink, R.C.; Caley, D.P.; Currie, J.M.; Brooks, S.A.; Carter, D.R. Over-expression of miR-31 or loss of KCNMA1 leads to increased cisplatin resistance in ovarian cancer cells. Tumor Biol. 2016, 37, 2565–2573. [Google Scholar] [CrossRef] [PubMed]
- Yang, W.; Wei, J.; Sun, T.; Liu, F. Effects of knockdown of miR-210 in combination with ionizing radiation on human hepatoma xenograft in nude mice. Radiat. Oncol. 2013, 8, 102. [Google Scholar] [CrossRef] [PubMed]
- Ng, W.L.; Yan, D.; Zhang, X.; Mo, Y.Y.; Wang, Y. Over-expression of miR-100 is responsible for the low-expression of ATM in the human glioma cell line: M059J. DNA Repair 2010, 9, 1170–1175. [Google Scholar] [CrossRef] [PubMed]
- Ye, C.; Sun, N.X.; Ma, Y.; Zhao, Q.; Zhang, Q.; Xu, C.; Wang, S.B.; Sun, S.H.; Wang, F.; Li, W. MicroRNA-145 contributes to enhancing radiosensitivity of cervical cancer cells. FEBS Lett. 2015, 589, 702–709. [Google Scholar] [CrossRef] [PubMed]
- Lv, L.; Li, Y.; Deng, H.; Zhang, C.; Pu, Y.; Qian, L.; Xiao, J.; Zhao, W.; Liu, Q.; Zhang, D.; et al. miR-193a-3p promotes the multi-chemoresistance of bladder cancer by targeting the HOXC9 gene. Cancer Lett. 2015, 357, 105–113. [Google Scholar] [CrossRef] [PubMed]
- Wouters, M.D.; van Gent, D.C.; Hoeijmakers, J.H.; Pothof, J. MicroRNAs, the DNA damage response and cancer. Mutat. Res. 2011, 717, 54–66. [Google Scholar] [CrossRef] [PubMed]
- Czochor, J.R.; Glazer, P.M. MicroRNAs in cancer cell response to ionizing radiation. Antioxid. Redox Signal. 2014, 21, 293–312. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Liu, G.L.; Liu, S.H.; Wang, C.X.; Xu, Y.L.; Ying, Y.; Mao, P. MicroRNA-148b enhances the radiosensitivity of non-hodgkin’s lymphoma cells by promoting radiation-induced apoptosis. J. Radiat. Res. 2012, 53, 516–525. [Google Scholar] [CrossRef] [PubMed]
- Wagner-Ecker, M.; Schwager, C.; Wirkner, U.; Abdollahi, A.; Huber, P.E. MicroRNA expression after ionizing radiation in human endothelial cells. Radiat. Oncol. 2010, 5, 25. [Google Scholar] [CrossRef] [PubMed]
- Gwak, H.S.; Kim, T.H.; Jo, G.H.; Kim, Y.J.; Kwak, H.J.; Kim, J.H.; Yin, J.; Yoo, H.; Lee, S.H.; Park, J.B. Silencing of microRNA-21 confers radio-sensitivity through inhibition of the PI3K/AKT pathway and enhancing autophagy in malignant glioma cell lines. PLoS ONE 2012, 7, e47449. [Google Scholar] [CrossRef] [PubMed]
- Chaudhry, M.A. Radiation-induced microRNA: Discovery, functional analysis, and cancer radiotherapy. J. Cell. Biochem. 2014, 115, 436–449. [Google Scholar] [CrossRef] [PubMed]
- Chaudhry, M.A.; Sachdeva, H.; Omaruddin, R.A. Radiation-induced microRNA modulation in glioblastoma cells differing in DNA-repair pathways. DNA Cell Biol. 2010, 29, 553–561. [Google Scholar] [CrossRef] [PubMed]
- Chaudhry, M.A. Real-time PCR analysis of micro-RNA expression in ionizing radiation-treated cells. Cancer Biother. Radiopharm. 2009, 24, 49–56. [Google Scholar] [CrossRef] [PubMed]
- Weidhaas, J.B.; Babar, I.; Nallur, S.M.; Trang, P.; Roush, S.; Boehm, M.; Gillespie, E.; Slack, F.J. MicroRNAs as potential agents to alter resistance to cytotoxic anticancer therapy. Cancer Res. 2007, 67, 11111–11116. [Google Scholar] [CrossRef] [PubMed]
- Saleh, A.D.; Savage, J.E.; Cao, L.; Soule, B.P.; Ly, D.; DeGraff, W.; Harris, C.C.; Mitchell, J.B.; Simone, N.L. Cellular stress induced alterations in microRNA let-7a and let-7b expression are dependent on p53. PLoS ONE 2011, 6, e24429. [Google Scholar] [CrossRef] [PubMed]
- Marta, G.N.; Garicochea, B.; Carvalho, A.L.; Real, J.M.; Kowalski, L.P. MicroRNAs, cancer and ionizing radiation: Where are we? Rev. Assoc. Med. Bras. 2015, 61, 275–281. [Google Scholar] [CrossRef] [PubMed]
- Novello, C.; Pazzaglia, L.; Conti, A.; Quattrini, I.; Pollino, S.; Perego, P.; Picci, P.; Benassi, M.S. p53-Dependent activation of microRNA-34a in response to etoposide-induced DNA damage in osteosarcoma cell lines not impaired by dominant negative p53 expression. PLoS ONE 2014, 9, e114757. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Zhu, H.; Yang, X.; Ge, Y.; Zhang, C.; Qin, Q.; Lu, J.; Zhan, L.; Cheng, H.; Sun, X. MicroRNA-21 is a novel promising target in cancer radiation therapy. Tumor Biol. 2014, 35, 3975–3979. [Google Scholar] [CrossRef] [PubMed]
- Metheetrairut, C.; Slack, F.J. MicroRNAs in the ionizing radiation response and in radiotherapy. Curr. Opin. Genet. Dev. 2013, 23, 12–19. [Google Scholar] [CrossRef] [PubMed]
- Daige, C.L.; Wiggins, J.F.; Priddy, L.; Nelligan-Davis, T.; Zhao, J.; Brown, D. Systemic delivery of a miR34a mimic as a potential therapeutic for liver cancer. Mol. Cancer Ther. 2014, 13, 2352–2360. [Google Scholar] [CrossRef] [PubMed]
- Conde, J.; Rosa, J.; de la Fuente, J.M.; Baptista, P.V. Gold-nanobeacons for simultaneous gene specific silencing and intracellular tracking of the silencing events. Biomaterials 2013, 34, 2516–2523. [Google Scholar] [CrossRef] [PubMed]
- Shah, M.Y.; Ferrajoli, A.; Sood, A.K.; Lopez-Berestein, G.; Calin, G.A. MicroRNA therapeutics in cancer—An emerging concept. EBioMedicine 2016, 12, 34–42. [Google Scholar] [CrossRef] [PubMed]

| Cancer | miRNA | Target(s) | Therapy | Effects | References |
|---|---|---|---|---|---|
| Breast | miR-18a | ATM | IR | Radiosensitivity | [21] |
| miR-155 | RAD51 | IR | Radiosensitivity | [37] | |
| miR-107 | RAD51 | PARP inhibitor | Chemosensitivity | [35] | |
| miR-182 | BRCA1 | IR | Radiosensitivity | [30] | |
| miR-34a | Bcl-2 | Docetaxel | Chemoresistance | [86] | |
| miR-21 | PTEN and PDCD4 | Chemotherapy | Chemoresistance | [87] | |
| miR-125b | BAK1 | Paclitaxel | Chemoresistance | [47] | |
| Lung | miR-138 | H2AX | IR | Radiosenstivity | [19] |
| miR-101 | ATM | IR | Radiosensitivity | [88] | |
| miR-497 | Bcl-2 | Chemotherapy | Chemosensitivity | [89] | |
| miR-34a | RAD51 | IR | Radiosensivity | [33] | |
| miR-155 | Apaf-1 | Chemotherapy | Chemorisistance | [64] | |
| miR-141 | PDCD4 | Cisplatin | Chemoresistance | [90] | |
| Ovarian | miR-506 | RAD51 | Chemotherapy | Chemosensitivity | [34] |
| miR-152 | ATG14 | Cisplatin | Chemosensitivity | [85] | |
| miR-31 | KCNMA1 | Cisplatin | Chemoresistance | [91] | |
| Hepatoma | miR-16 | Bcl-2 | Epigallocatechin gallate | Chemosensitivity | [61] |
| miR-210 | AIFM3 | IR | Radioresistance | [92] | |
| Colorectal | miR-145 | RAD18 | 5-FU | Chemosensitivity | [32] |
| miR-203 | ATM | Oxaliplatin | Chemoresistance | [24] | |
| Glioblastoma | miR-100 | ATM | IR | Radioresistance | [93] |
| miR-338-5P | RHEB | IR | Radiosensitivity | [78] | |
| miR-181b | SENP2 | IR | Radioresistance | [87] | |
| Renal carcinoma | miR-185 | ATR | IR | Radiosensitivity | [27] |
| Prostate | miR-744-3P | RAD23B | IR | Radiosensitivity | [42] |
| miR-890 | MAD2L2, WEE1, XPC | IR | Radiosensitivity | [42] | |
| Cervical | miR-145 | HLTF | IR | Radiosensitivity | [94] |
| Bladder | miR-193a-3P | HOXC9 | Chemotherapy | Chemoresistance | [95] |
| Hematopoietic | miR-24 | H2AX | Cisplatin | Chemosensitivity | [96] |
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
He, M.; Zhou, W.; Li, C.; Guo, M. MicroRNAs, DNA Damage Response, and Cancer Treatment. Int. J. Mol. Sci. 2016, 17, 2087. https://doi.org/10.3390/ijms17122087
He M, Zhou W, Li C, Guo M. MicroRNAs, DNA Damage Response, and Cancer Treatment. International Journal of Molecular Sciences. 2016; 17(12):2087. https://doi.org/10.3390/ijms17122087
Chicago/Turabian StyleHe, Mingyang, Weiwei Zhou, Chuang Li, and Mingxiong Guo. 2016. "MicroRNAs, DNA Damage Response, and Cancer Treatment" International Journal of Molecular Sciences 17, no. 12: 2087. https://doi.org/10.3390/ijms17122087
APA StyleHe, M., Zhou, W., Li, C., & Guo, M. (2016). MicroRNAs, DNA Damage Response, and Cancer Treatment. International Journal of Molecular Sciences, 17(12), 2087. https://doi.org/10.3390/ijms17122087