Pegylated Trastuzumab Fragments Acquire an Increased in Vivo Stability but Show a Largely Reduced Affinity for the Target Antigen
Abstract
:1. Introduction
2. Results
2.1. Preparation of TrastFab
2.4. Preparation of di-PEGylated TrastFab’
2.5. ELISA Binding Studies
2.6. Surface Plasmon Resonance (SPR) Binding Measurements
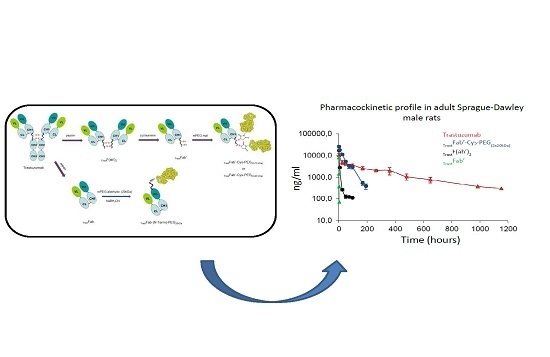
2.7. Pharmacokinetic Properties
3. Discussion
4. Materials and Methods
4.1. Media and Reagents
4.2. TrastFab Preparation
4.3. TrastF(ab’)2 Preparation
4.4. Preparation of the Fab’ of Trastuzumab
4.5. Preparation of Trastuzumab Fab’ Di-PEGylated on the Hinge Cysteine Residues
4.6. N-terminal Conjugation of TrastFab
4.7. SDS-PAGE Analysis
4.8. Western Blotting Analysis of TrastFab-(N-Term)-PEG20 kDa and TrastFab’-Cys-PEG(2 × 10 kDa)
4.9. Analysis of Affinity by ELISA
4.10. Statistical Analysis
4.11. Analysis of Binding by SPR
4.12. Pharmacokinetic Studies
5. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Carter, P.; Presta, L.; Gorman, C.M.; Ridgway, J.B.; Henner, D.; Wong, W.L.; Rowland, A.M.; Kotts, C.; Carver, M.E.; Shepard, H.M. Humanization of an anti-p185HER2 antibody for human cancer therapy. Proc. Natl. Acad. Sci. USA 1992, 89, 4285–4289. [Google Scholar] [CrossRef] [PubMed]
- Fendly, B.M.; Winget, M.; Hudziak, R.M.; Lipari, M.T.; Napier, M.A.; Ullrich, A. Characterization of murine monoclonal antibodies reactive to either the human epidermal growth factor receptor or HER2/neu gene product. Cancer Res. 1990, 50, 1550–1558. [Google Scholar] [PubMed]
- Slamon, D.J.; Leyland-Jones, B.; Shak, S.; Fuchs, H.; Paton, V.; Bajamonde, A.; Fleming, T.; Eiermann, W.; Wolter, J.; Pegram, M.; et al. Use of chemotherapy plus a monoclonal antibody against HER2 for metastatic breast cancer that overexpresses HER2. N. Engl. J. Med. 2001, 344, 783–792. [Google Scholar] [CrossRef] [PubMed]
- Cobleigh, M.A.; Vogel, C.L.; Tripathy, D.; Robert, N.J.; Scholl, S.; Fehrenbacher, L.; Wolter, J.M.; Paton, V.; Shak, S.; Lieberman, G.; et al. Multinational study of the efficacy and safety of humanized anti-HER2 monoclonal antibody in women who have HER2-overexpressing metastatic breast cancer that has progressed after chemotherapy for metastatic disease. J. Clin. Oncol. 1999, 17, 2639–2648. [Google Scholar] [PubMed]
- Romond, E.H.; Perez, E.A.; Bryant, J.; Suman, V.J.; Geyer, C.E., Jr.; Davidson, N.E.; Tan-Chiu, E.; Martino, S.; Paik, S.; Kaufman, P.A.; et al. Trastuzumab plus adjuvant chemotherapy for operable HER2-positive breast cancer. N. Engl. J. Med. 2005, 353, 1673–1684. [Google Scholar] [CrossRef] [PubMed]
- Holliger, P.; Hudson, P.J. Engineered antibody fragments and the rise of single domains. Nat. Biotechnol. 2005, 23, 1126–1136. [Google Scholar] [CrossRef] [PubMed]
- Kourlas, H.; Abrams, P. Ranibizumab for the treatment of neovascular age-related macular degeneration: A review. Clin. Ther. 2007, 29, 1850–1861. [Google Scholar] [CrossRef] [PubMed]
- Fischer, N.; Leger, O. Bispecific antibodies: Molecules that enable novel therapeutic strategies. Pathobiology 2007, 74, 3–14. [Google Scholar] [CrossRef] [PubMed]
- Spiegelberg, H.L.; Grey, H.M. Catabolism of human γG immunoglobulins of different heavy chain subclasses: II, catabolism of γG myeloma proteins in heterologous species. J. Immunol. 1968, 101, 711–716. [Google Scholar] [CrossRef] [PubMed]
- Bruno, R.; Washington, C.B.; Lu, J.F.; Lieberman, G.; Banken, L.; Klein, P. Population pharmacokinetics of trastuzumab in patients with HER2+ metastatic breast cancer. Cancer Chemother. Pharmacol. 2005, 56, 361–369. [Google Scholar] [CrossRef] [PubMed]
- Beckman, R.A.; Weiner, L.M.; Davis, H.M. Antibody constructs in cancer therapy: Protein engineering strategies to improve exposure in solid tumors. Cancer 2007, 109, 170–179. [Google Scholar] [CrossRef] [PubMed]
- Pasut, G.; Veronese, F.M. PEGylation for improving the effectiveness of therapeutic biomolecules. Drugs Today (Barc.) 2009, 45, 687–695. [Google Scholar] [CrossRef] [PubMed]
- Veronese, F.M.; Pasut, G. PEGylation, successfull approach to drug delivery. Drug Discov. Today 2005, 10, 1451–1458. [Google Scholar] [CrossRef]
- Roberts, M.J.; Bentley, M.D.; Harris, J.M. Chemistry for peptide and protein PEGylation. Drug Deliv. Rev. 2002, 54, 459–447. [Google Scholar] [CrossRef]
- Khalili, H.; Godwin, A.; Choi, J.W.; Lever, R.; Brocchini, S. Comparative binding of disulfide-bridged PEG-Fabs. Bioconjug. Chem. 2012, 23, 2262–2277. [Google Scholar] [CrossRef] [PubMed]
- Harlow, E.; Lane, D. Antibodies: A Laboratory Manual; Cold Spring Harbor Laboratory: Cold Spring Harbor, NY, USA, 1988; pp. 628–629. [Google Scholar]
- Kurfurst, M.M. Detection and molecular-weight determination of polyethylene glycol-modified hirudin by staining after sodium dodecyl-sulfate polyacrylamide-gel electrophoresis. Anal. Biochem. 1992, 200, 244–248. [Google Scholar] [CrossRef]
- Tong, Y.; Zhong, K.; Tian, H.; Gao, X.D.; Xu, X.Y.; Yin, X.J.; Yao, W.B. Characterization of a monoPEG20000-Endostar. Int. J. Biol. Macromol. 2010, 46, 331–336. [Google Scholar] [CrossRef] [PubMed]
- Zheng, C.; Ma, G.; Su, Z. Native PAGE eliminates the problem of PEG-SDS interaction in SDS-PAGE and provides an alternative to HPLC in characterization of protein PEGylation. Electrophoresis 2007, 28, 2801–2807. [Google Scholar] [CrossRef] [PubMed]
- Fee, C.J.; van Alstine, J.M. Prediction of the Viscosity Radius and the Size Exclusion Chromatography Behavior of PEGylated Proteins. Bioconjug. Chem. 2004, 15, 1304–1313. [Google Scholar] [CrossRef] [PubMed]
- Hu, J.; Sebald, W. N-terminal specificity of PEGylation of human bone morphogenetic protein-2 at acidic pH. Int. J. Pharm. 2011, 413, 140–146. [Google Scholar] [CrossRef] [PubMed]
- Seely, J.E.; Richey, C.W. Use of ion-exchange chromatography and hydrophobic interaction chromatography in the preparation and recovery of polyethylene glycol-linked proteins. J. Chromatogr. A 2001, 908, 235–241. [Google Scholar] [CrossRef]
- Pepinsky, R.B.; Walus, L.; Shao, Z.; Ji, B.; Gu, S.; Sun, Y.; Wen, D.; Lee, X.; Wang, Q.; Garber, E.; et al. Production of a PEGylated Fab’ of the anti-LINGO-1 Li33 antibodyand assessment of its biochemical and functional properties in vitro and in a rat model of remyelination. Bioconjug. Chem. 2011, 22, 200–210. [Google Scholar] [CrossRef] [PubMed]
- Kubetzko, S.; Sarkar, C.A.; Plückthun, A. Protein PEGylation decreases observed target association rates via a dual blocking mechanism. Mol. Pharmacol. 2005, 68, 1439–1454. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, M.; Kato, C.; Kato, A. Therapeutic antibodies: Their mechanisms of action and the pathological findings they induce in toxicity studies. J. Toxicol. Pathol. 2015, 28, 133–139. [Google Scholar] [CrossRef] [PubMed]
- Ginn, C.; Khalili, H.; Lever, R.; Brocchini, S. PEGylation and its impact on the design of new protein-based medicines. Future Med. Chem. 2014, 6, 1829–1846. [Google Scholar] [CrossRef] [PubMed]
- Molineux, G. Pegfilgrastim: Using pegylation technology to improve neutropenia support in cancer patients. Anticancer Drugs 2003, 14, 259–264. [Google Scholar] [CrossRef] [PubMed]
- Olafsen, T.; Wu, A.M. Antibody vectors for imaging. Semin. Nucl. Med. 2010, 40, 167–181. [Google Scholar] [CrossRef] [PubMed]
- Freise, A.C.; Wu, A.M. In vivo imaging with antibodies and engineered fragments. Mol. Immunol. 2015, 67, 142–152. [Google Scholar] [CrossRef] [PubMed]
- Seldon, T.A.; Hughes, K.E.; Munster, D.J.; Chin, D.Y.; Jones, M.L. Improved Protein-A separation of VH3 Fab from Fc after papain digestion of antibodies. J. Biomol. Tech. 2011, 22, 50–52. [Google Scholar] [PubMed]
- Fasman, D.G. Practical Handbook of Biochemistry and Molecular Biology; CRC Press: Boston, MA, USA, 1992. [Google Scholar]
- Mariani, M.; Cauragra, M.; Tarditi, L.; Seccariani, E. A new menzymatic method to obtain high-yield F(ab)2 suitable for clinical use from mouse IgG1. Mol. Immunol. 1991, 28, 69–77. [Google Scholar] [CrossRef]
- Andrew, S.M.; Titus, J.A. Current Protocols in Immunology; Wiley: New York, NY, USA, 1997; pp. 2.8.1–2.8.10. [Google Scholar]
- Humphreys, D.P.; Heywood, S.P.; Henry, A.; Ait-Lhadj, L.; Antoniw, P.; Palframan, R.; Greenslade, K.J.; Carrington, B.; Reeks, D.G.; Bowering, L.C.; et al. Alternative antibody Fab’ fragment PEGylation strategies: Combination of strong reducing agents, disruption of the interchain disulphide bond and disulphide engineering. Protein Eng. Des. Sel. 2007, 20, 227–234. [Google Scholar] [CrossRef] [PubMed]
- Laemmli, U.K. Cleavage of structural proteins during assembly of the head of bacteriophage T4. Nature 1970, 227, 680–685. [Google Scholar] [CrossRef] [PubMed]
- Johnsson, B.; Lofas, S.; Lindquist, G. Immobilization of proteins to a carboxymethyldextran-modified gold surface for biospecific interaction analysis in surface plasmon resonance sensors. Anal. Biochem. 1991, 198, 268–277. [Google Scholar] [CrossRef]
- Sandomenico, A.; Severino, V.; Chambery, A.; Focà, A.; Focà, G.; Farina, C.; Ruvo, M. A comparative structural and bioanalytical study of IVIG clinical lots. Mol. Biotechnol. 2013, 54, 983–995. [Google Scholar] [CrossRef] [PubMed]




| Product Number | Product Description | Predicted Mw (kDa) | Determined Mw (SE-HPLC) (kDa) | Calculated Molecular Volume (nm3) # |
|---|---|---|---|---|
| 1 | Trastuzumab | 150 | 150 | 434 |
| 2 | TrastFab | 50 | 36 * | 163 |
| 3 | TrastF(ab’)2 | 100 | 36 * | 288 |
| 4 | TrastFab’ | 50 | 101 | 163 |
| 5 | TrastFab’-Cys-PEG(2 × 10 kDa) | 70 | 490 | 700 |
| 6 | TrastFab’-Cys-PEG(2 × 20 kDa) | 90 | 470 | 2000 |
| 7 | TrastFab-(N-Term)-PEG20 kDa | 90 ** | 771 | 700 |
| Product | EC50 (nM) |
|---|---|
| Trastuzumab | 0.228 ± 0.06 |
| TrastFab | 0.459 ± 0.06 |
| TrastFab’ | 0.336 ± 0.08 |
| TrastF(ab’)2 | 0.238 ± 0.03 |
| TrastFab-(N-Term)-PEG20 kDa | 0.839 ± 0.02 |
| TrastFab’-Cys-PEG(2 × 10 kDa) | 0.903 ± 0.13 |
| TrastFab’-Cys-PEG(2 × 20 kDa) | 0.697 ± 0.14 |
| Product | Ka (×104) M−1·s−1 | Kd (×10−5) s−1 | KD (kd/ka) nM |
|---|---|---|---|
| Trastuzumab | 62.3 | 8.16 | 0.131 |
| TrastFab | 59.9 | 24.2 | 0.405 |
| TrastFab’ | 57.9 | 17.2 | 0.297 |
| TrastF(ab’)2 | 32.8 | 6.17 | 0.188 |
| TrastFab-(N-Term)-PEG20 kDa | 7.71 | 37.2 | 4.82 |
| TrastFab’-Cys-PEG(2 × 10 kDa) | 34.5 | 28.5 | 0.827 |
| TrastFab’-Cys-PEG(2 × 20 kDa) | 7.03 | 15.8 | 2.24 |
| Product | Dose | C max (ng/mL) | AUC (ng/mL h) | t1/2 α (h) | t1/2 β (h) |
|---|---|---|---|---|---|
| Trastuzumab | 2.0 mg/kg | 26,315.8 * | 1,694,759.1 (1152 h) | 22.1 | 288.8 |
| TrastF(ab’)2 | 2.0 mg/kg | 18,947.4 * | 116,293.6 (96 h) | 4.5 | 48.1 |
| TrastFab’ | 2.0 mg/kg | 25,0002 + | 4985.3 (2 h) | 0.13 | 0.36 |
| TrastFab’-Cys-PEG(2 × 20 kDa) | 2.0 mg/kg | 26,315.8 * | 896,387.8 (192 h) | 11 | 35.4 |
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons by Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Selis, F.; Focà, G.; Sandomenico, A.; Marra, C.; Di Mauro, C.; Saccani Jotti, G.; Scaramuzza, S.; Politano, A.; Sanna, R.; Ruvo, M.; et al. Pegylated Trastuzumab Fragments Acquire an Increased in Vivo Stability but Show a Largely Reduced Affinity for the Target Antigen. Int. J. Mol. Sci. 2016, 17, 491. https://doi.org/10.3390/ijms17040491
Selis F, Focà G, Sandomenico A, Marra C, Di Mauro C, Saccani Jotti G, Scaramuzza S, Politano A, Sanna R, Ruvo M, et al. Pegylated Trastuzumab Fragments Acquire an Increased in Vivo Stability but Show a Largely Reduced Affinity for the Target Antigen. International Journal of Molecular Sciences. 2016; 17(4):491. https://doi.org/10.3390/ijms17040491
Chicago/Turabian StyleSelis, Fabio, Giuseppina Focà, Annamaria Sandomenico, Carla Marra, Concetta Di Mauro, Gloria Saccani Jotti, Silvia Scaramuzza, Annalisa Politano, Riccardo Sanna, Menotti Ruvo, and et al. 2016. "Pegylated Trastuzumab Fragments Acquire an Increased in Vivo Stability but Show a Largely Reduced Affinity for the Target Antigen" International Journal of Molecular Sciences 17, no. 4: 491. https://doi.org/10.3390/ijms17040491
APA StyleSelis, F., Focà, G., Sandomenico, A., Marra, C., Di Mauro, C., Saccani Jotti, G., Scaramuzza, S., Politano, A., Sanna, R., Ruvo, M., & Tonon, G. (2016). Pegylated Trastuzumab Fragments Acquire an Increased in Vivo Stability but Show a Largely Reduced Affinity for the Target Antigen. International Journal of Molecular Sciences, 17(4), 491. https://doi.org/10.3390/ijms17040491