Changes in the Arabidopsis thaliana Proteome Implicate cAMP in Biotic and Abiotic Stress Responses and Changes in Energy Metabolism
Abstract
:1. Introduction
2. Results and Discussion
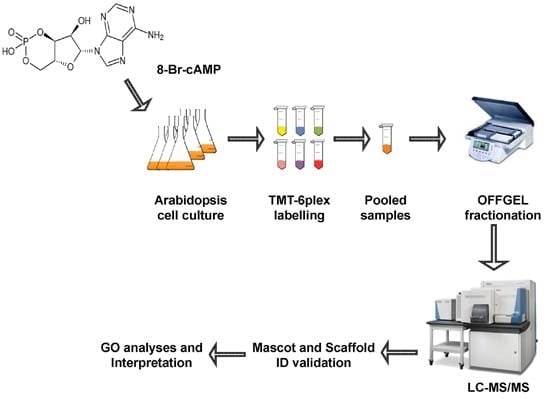
3. Materials and Methods
3.1. Plant Material and Growth Conditions
3.2. cAMP Treatment and Protein Extraction
3.3. Peptide Labeling Using Tandem Mass Tag and Peptide Fractionation by OFFGEL Fractionator
3.4. Protein Identification by LTQ Orbitrap and Quantification of Differentially Expressed Proteins
3.5. Computational Analysis of Functional Enrichment
4. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Gancedo, J.M. Biological roles of cAMP: Variations on a theme in the different kingdoms of life. Biol. Rev. 2013, 88, 645–668. [Google Scholar] [CrossRef] [PubMed]
- Moutinho, A.; Hussey, P.; Trewavas, A.; Malhó, R. Cyclic AMP acts as a second messenger in pollen tube growth and reorientation. Proc. Natl. Acad. Sci. USA 2001, 98, 10481–10486. [Google Scholar] [CrossRef] [PubMed]
- Cooke, C.J.; Smith, C.J.; Walton, T.J.; Newton, R.P. Evidence that cyclic AMP is involved in the hypersensitive response of Medicago sativa to a fungal elicitor. Phytochemistry 1994, 35, 889–895. [Google Scholar] [CrossRef]
- Kim, H.-S.; Park, S.-Y.; Lee, S.; Adams, E.L.; Czymmek, K.; Kang, S. Loss of cAMP-dependent protein kinase A affects multiple traits important for root pathogenesis by Fusarium oxysporum. Mol. Plant Microbe Interact. 2011, 24, 719–732. [Google Scholar] [CrossRef] [PubMed]
- Lemtiri-Chlieh, F.; Thomas, L.; Marondedze, C.; Irving, H.; Gehring, C. Cyclic nucleotides and nucleotide cyclases in Plant stress responses. In Abiotic Stress Response in Plants—Physiological, Biochemical and Genetic Perspectives; Shanker, A., Venkateswarlu, B., Eds.; InTech: Vienna, Austria, 2011; p. 346p. [Google Scholar]
- Ma, W.; Qi, Z.; Smigel, A.; Walker, R.; Verma, R.; Berkowitz, G. Ca2+, cAMP, and transduction of non-self perception during plant immune responses. Proc. Natl. Acad. Sci. USA 2009, 106, 20995–21000. [Google Scholar] [CrossRef] [PubMed]
- Lemtiri-Chlieh, F.; Berkowitz, G.A. Cyclic adenosine monophosphate regulates calcium channels in the plasma membrane of Arabidopsis leaf guard and mesophyll cells. J. Biol. Chem. 2004, 279, 35306–35312. [Google Scholar] [CrossRef] [PubMed]
- Qi, Z.; Verma, R.; Gehring, C.; Yamaguchi, Y.; Zhao, Y.; Ryan, C.; Berkowitz, G. Ca2+ signaling by plant Arabidopsis thaliana Pep peptides depends on AtPepR1, a receptor with guanylyl cyclase activity, and cGMP-activated Ca2+ channels. Proc. Natl. Acad. Sci. USA 2010, 107, 21193–21198. [Google Scholar] [CrossRef] [PubMed]
- Kugler, A.; Köhler, B.; Palme, K.; Wolff, P.; Dietrich, P. Salt-dependent regulation of a CNG channel subfamily in Arabidopsis. BMC Plant Biol. 2009, 9, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Kaplan, B.; Sherman, T.; Fromm, H. Cyclic nucleotide-gated channels in plants. FEBS Lett. 2007, 581, 2237–2246. [Google Scholar] [CrossRef] [PubMed]
- Ladwig, F.; Dahlke, R.I.; Stührwohldt, N.; Hartmann, J.; Harter, K.; Sauter, M. Phytosulfokine regulates growth in Arabidopsis through a response module at the plasma membrane that includes CYCLIC NUCLEOTIDE-GATED CHANNEL17, H+-ATPase, and BAK1. Plant Cell 2015, 27, 1718–1729. [Google Scholar] [CrossRef] [PubMed]
- DeWald, D.B.; Torabinejad, J.; Jones, C.A.; Shope, J.C.; Cangelosi, A.R.; Thompson, J.E.; Prestwich, G.D.; Hama, H. Rapid Accumulation of Phosphatidylinositol 4,5-Bisphosphate and Inositol 1,4,5-Trisphosphate Correlates with Calcium Mobilization in Salt-Stressed Arabidopsis. Plant Physiol. 2001, 126, 759–769. [Google Scholar] [CrossRef] [PubMed]
- Frietsch, S.; Wang, Y.-F.; Sladek, C.; Poulsen, L.R.; Romanowsky, S.M.; Schroeder, J.I.; Harper, J.F. A cyclic nucleotide-gated channel is essential for polarized tip growth of pollen. Proc. Natl. Acad. Sci. USA 2007, 104, 14531–14536. [Google Scholar] [CrossRef] [PubMed]
- Konrad, K.R.; Hedrich, R. The use of voltage-sensitive dyes to monitor signal-induced changes in membrane potential–ABA triggered membrane depolarization in guard cells. Plant J. 2008, 55, 161–173. [Google Scholar] [CrossRef] [PubMed]
- Ma, W.; Berkowitz, G.A. Ca2+ conduction by plant cyclic nucleotide gated channels and associated signaling components in pathogen defense signal transduction cascades. New Phytol. 2011, 190, 566–572. [Google Scholar] [CrossRef] [PubMed]
- Maathuis, F.J.M.; Sanders, D. Sodium uptake in Arabidopsis roots is regulated by cyclic nucleotides. Plant Physiol. 2001, 127, 1617–1625. [Google Scholar] [CrossRef] [PubMed]
- Gehring, C. Adenyl cyclases and cAMP in plant signaling—Past and present. Cell Commun. Signal. 2010, 8, 15–19. [Google Scholar] [CrossRef] [PubMed]
- Marondedze, C.; Wong, A.; Thomas, L.; Irving, H.; Gehring, C. Cyclic Nucleotide Monophosphates in Plants and Plant Signaling. In Handbook of Experimental Pharmacology; Barrett, J.E., Flockerzi, V., Frohman, M.A., Geppetti, P., Hofmann, F.B., Michel, M.C., Page, C.P., Thorburn, A.M., Wang, K., Eds.; Springer: Berlin, Germany; Heidelberg, Germany, 2016; pp. 1–17. [Google Scholar]
- Al-Younis, I.; Wong, A.; Gehring, C. The Arabidopsis thaliana K+-uptake permease 7 (AtKUP7) contains a functional cytosolic adenylate cyclase catalytic centre. FEBS Lett. 2015, 589, 3848–3852. [Google Scholar] [CrossRef] [PubMed]
- Newton, R.; Gibbs, N.; Moyse, C.; Wiebers, J.; Brown, E. Mass spectrometric identification of adenosine 3′:5′-cyclic monophosphate isolated from a higher plant tissue. Phytochemistry 1980, 19, 1909–1911. [Google Scholar] [CrossRef]
- Donaldson, L.; Meier, S.; Gehring, C. The Arabidopsis cyclic nucleotide interactome. Cell Commun. Signal. 2016, 14, 10–27. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thomas, L.; Marondedze, C.; Ederli, L.; Pasqualini, S.; Gehring, C. Proteomic signatures implicate cAMP in light and temperature responses in Arabidopsis thaliana. J. Proteom. 2013, 83, 47–59. [Google Scholar] [CrossRef] [PubMed]
- Meier, S.; Gehring, C. A guide to the integrated application of on-line data mining tools for the inference of gene functions at the systems level. Biotechnol. J. 2008, 3, 1375–1387. [Google Scholar] [CrossRef] [PubMed]
- Alqurashi, M.; Meier, S. Inferring biological functions of guanylyl cyclases with computational methods. In Cyclic Nucleotide Signaling in Plants; Gehring, C., Ed.; Humana Press: Totowa, NJ, USA, 2013; Volume 1016, pp. 225–234. [Google Scholar]
- Flechner, A.; Gross, W.; Martin, W.F.; Schnarrenberger, C. Chloroplast class I and class II aldolases are bifunctional for fructose-1,6-biphosphate and sedoheptulose-1,7-biphosphate cleavage in the Calvin cycle. FEBS Lett. 1999, 447, 200–202. [Google Scholar] [CrossRef]
- Lu, W.; Tang, X.; Huo, Y.; Xu, R.; Qi, S.; Huang, J.; Zheng, C.; Wu, C. Identification and characterization of fructose 1,6-bisphosphate aldolase genes in Arabidopsis reveal a gene family with diverse responses to abiotic stresses. Gene 2012, 503, 65–74. [Google Scholar] [CrossRef] [PubMed]
- Nunes-Nesi, A.; Carrari, F.; Lytovchenko, A.; Smith, A.M.O.; Ehlers Loureiro, M.; Ratcliffe, R.G.; Sweetlove, L.J.; Fernie, A.R. Enhanced photosynthetic performance and growth as a consequence of decreasing mitochondrial malate dehydrogenase activity in transgenic tomato plants. Plant Physiol. 2005, 137, 611–622. [Google Scholar] [CrossRef] [PubMed]
- Araújo, W.L.; Nunes-Nesi, A.; Nikoloski, Z.; Sweetlove, L.J.; Fernie, A.R. Metabolic control and regulation of the tricarboxylic acid cycle in photosynthetic and heterotrophic plant tissues. Plant Cell Environ. 2012, 35, 1–21. [Google Scholar] [CrossRef] [PubMed]
- Scheibe, R. Malate valves to balance cellular energy supply. Physiol. Plant. 2004, 120, 21–26. [Google Scholar] [CrossRef] [PubMed]
- Taylor, N.L.; Heazlewood, J.L.; Day, D.A.; Millar, A.H. Differential impact of environmental stresses on the pea mitochondrial proteome. Mol. Cell. Proteom. 2005, 4, 1122–1133. [Google Scholar] [CrossRef] [PubMed]
- Komatsu, S.; Yamamoto, A.; Nakamura, T.; Nouri, M.-Z.; Nanjo, Y.; Nishizawa, K.; Furukawa, K. Comprehensive analysis of mitochondria in roots and hypocotyls of soybean under flooding stress using proteomics and metabolomics techniques. J. Proteome Res. 2011, 10, 3993–4004. [Google Scholar] [CrossRef] [PubMed]
- Nunes-Nesi, A.; Araújo, W.L.; Fernie, A.R. Targeting mitochondrial metabolism and machinery as a means to enhance photosynthesis. Plant Physiol. 2011, 155, 101–107. [Google Scholar] [CrossRef] [PubMed]
- Tronconi, M.A.; Fahnenstich, H.; Gerrard Weehler, M.C.; Andreo, C.S.; Flügge, U.-I.; Drincovich, M.F.; Maurino, V.G. Arabidopsis NAD-malic enzyme functions as a homodimer and heterodimer and has a major impact on nocturnal metabolism. Plant Physiol. 2008, 146, 1540–1552. [Google Scholar] [CrossRef] [PubMed]
- Kovacs, D.; Kalmar, E.; Torok, Z.; Tompa, P. Chaperone activity of ERD10 and ERD14, two disordered stress-related plant proteins. Plant Physiol. 2008, 147, 381–390. [Google Scholar] [CrossRef] [PubMed]
- Thornalley, P.J. Glyoxalase I—Structure, function and a critical role in the enzymatic defence against glycation. Biochem. Soc. Trans. 2003, 31, 1343–1348. [Google Scholar] [CrossRef] [PubMed]
- Mustafiz, A.; Singh, A.; Pareek, A.; Sopory, S.; Singla-Pareek, S. Genome-wide analysis of rice and Arabidopsis identifies two glyoxalase genes that are highly expressed in abiotic stresses. Funct. Integr. Genom. 2011, 11, 293–305. [Google Scholar] [CrossRef] [PubMed]
- Finkelstein, R.R.; Rock, C.D. Abscisic acid biosynthesis and response. Arab. Book 2002, 1, e0058. [Google Scholar] [CrossRef] [PubMed]
- Cutler, S.R.; Rodriguez, P.L.; Finkelstein, R.R.; Abrams, S.R. Abscisic acid: Emergence of a core signaling network. Annu. Rev. Plant Biol. 2010, 61, 651–679. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.-K. Salt and drought stress signal transduction in plants. Annu. Rev. Plant Biol. 2002, 53, 247–273. [Google Scholar] [CrossRef] [PubMed]
- Harwood, J.L. Fatty acid metabolism. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1988, 39, 101–138. [Google Scholar] [CrossRef]
- Leonhardt, N.; Kwak, J.M.; Robert, N.; Waner, D.; Leonhardt, G.; Schroeder, J.I. Microarray expression analyses of Arabidopsis guard cells and isolation of a recessive abscisic acid hypersensitive protein phosphatase 2C mutant. Plant Cell 2004, 16, 596–615. [Google Scholar] [CrossRef] [PubMed]
- Trupkin, S.A.; Mora-García, S.; Casal, J.J. The cyclophilin ROC1 links phytochrome and cryptochrome to brassinosteroid sensitivity. Plant J. 2012, 71, 712–723. [Google Scholar] [CrossRef] [PubMed]
- Matsui, A.; Ishida, J.; Morosawa, T.; Mochizuki, Y.; Kaminuma, E.; Endo, T.A.; Okamoto, M.; Nambara, E.; Nakajima, M.; Kawashima, M.; et al. Arabidopsis transcriptome analysis under drought, cold, high-salinity and ABA treatment conditions using a tiling array. Plant Cell Physiol. 2008, 49, 1135–1149. [Google Scholar] [CrossRef] [PubMed]
- Kuhn, J.M.; Schroeder, J.I. Impacts of altered RNA metabolism on abscisic acid signaling. Curr. Opin. Plant Biol. 2003, 6, 463–469. [Google Scholar] [CrossRef]
- Ghelis, T.; Bolbach, G.; Clodic, G.; Habricot, Y.; Miginiac, E.; Sotta, B.; Jeannette, E. Protein tyrosine kinases and protein tyrosine phosphatases are involved in abscisic acid-dependent processes in Arabidopsis seeds and suspension cells. Plant Physiol. 2008, 148, 1668–1680. [Google Scholar] [CrossRef] [PubMed]
- Gomez, J.; Sanchez-Martinez, D.; Stiefel, V.; Rigau, J.; Puigdomenech, P.; Pages, M. A gene induced by the plant hormone abscisic acid in response to water stress encodes a glycine-rich protein. Nature 1988, 334, 262–264. [Google Scholar] [CrossRef] [PubMed]
- Testi, M.G.; Croce, R.; Laureto, P.P.-D.; Bassi, R. A CK2 site is reversibly phosphorylated in the photosystem II subunit CP29. FEBS Lett. 1996, 399, 245–250. [Google Scholar] [CrossRef]
- Amme, S.; Matros, A.; Schlesier, B.; Mock, H.-P. Proteome analysis of cold stress response in Arabidopsis thaliana using DIGE-technology. J. Exp. Bot. 2006, 57, 1537–1546. [Google Scholar] [CrossRef] [PubMed]
- Mauch-Mani, B.; Mauch, F. The role of abscisic acid in plant–pathogen interactions. Curr. Opin. Plant Biol. 2005, 8, 409–414. [Google Scholar] [CrossRef] [PubMed]
- Mohr, P.G.; Cahill, D.M. Abscisic acid influences the susceptibility of Arabidopsis thaliana to Pseudomonas syringae pv. tomato and Peronospora parasitica. Funct. Plant Biol. 2003, 30, 461–469. [Google Scholar] [CrossRef]
- Timmusk, S.; Wagner, E.G.H. The plant-growth-promoting rhizobacterium Paenibacillus polymyxa induces changes in Arabidopsis thaliana gene expression: A possible connection between biotic and abiotic stress responses. Mol. Plant-Microbe Interact. 1999, 12, 951–959. [Google Scholar] [CrossRef] [PubMed]
- Kariola, T.; Brader, G.; Helenius, E.; Li, J.; Heino, P.; Palva, E.T. Early Response to Dehydration 15. A negative Regulator of ABA-responses in Arabidopsis. Plant Physiol. 2006, 142, 1559–1573. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.; Chao, G.; Singh, K.B. The promoter of a H2O2-inducible, Arabidopsis glutathione S-transferase gene contains closely linked OBF- and OBP1-binding sites. Plant J. 1996, 10, 955–966. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.; Singh, K.B. The auxin, hydrogen peroxide and salicylic acid induced expression of the Arabidopsis GST6 promoter is mediated in part by an ocs element. Plant J. 1999, 19, 667–677. [Google Scholar] [CrossRef] [PubMed]
- Wagner, U.; Edwards, R.; Dixon, D.; Mauch, F. Probing the diversity of the Arabidopsis glutathione S-transferase gene family. Plant Mol. Biol. 2002, 49, 515–532. [Google Scholar] [CrossRef] [PubMed]
- Uquillas, C.; Letelier, I.; Blanco, F.; Jordana, X.; Holuigue, L. NPR1-Independent Activation of Immediate Early Salicylic Acid-Responsive Genes in Arabidopsis. Mol. Plant Microbe Interact. 2004, 17, 34–42. [Google Scholar] [CrossRef] [PubMed]
- Perl-Treves, R.; Foley, R.C.; Chen, W.; Singh, K.B. Early Induction of the Arabidopsis GSTF8 Promoter by Specific Strains of the Fungal Pathogen Rhizoctonia solani. Mol. Plant Microbe Interact. 2004, 17, 70–80. [Google Scholar] [CrossRef] [PubMed]
- Kimura, M.; Yamamoto, Y.Y.; Seki, M.; Sakurai, T.; Sato, M.; Abe, T.; Yoshida, S.; Manabe, K.; Shinozaki, K.; Matsui, M. Identification of Arabidopsis genes regulated by high light-stress using cDNA microarray. Photochem. Photobiol. 2003, 77, 226–233. [Google Scholar] [CrossRef]
- Smýkalová, I.; Zámečníková, B. The relationship between salinity and cadmium stress in barley. Biol. Plant. 2003, 46, 269–273. [Google Scholar] [CrossRef]
- Roth, U.; von Roepenack-Lahaye, E.; Clemens, S. Proteome changes in Arabidopsis thaliana roots upon exposure to Cd2+. J. Exp. Bot. 2006, 57, 4003–4013. [Google Scholar] [CrossRef] [PubMed]
- Pietrowska-Borek, M.; Nuc, K.; Chadzinikolau, T.; Lechowska, K. Cadmium stress causes accumulation of cAMP and this signal molecule induces phenylpropanoid pathway in Arabidopsis thaliana seedlings. In Proceedings of the First Polish-German Biochemical Societies Joint Meeting, Poznon, Poland, 11–14 September 2012. poster 4.30.
- Pietrowska-Borek, M.; Nuc, K. Both cyclic-AMP and cyclic-GMP can act as regulators of the phenylpropanoid pathway in Arabidopsis thaliana seedlings. Plant Physiol. Biochem. 2013, 70, 142–149. [Google Scholar] [CrossRef] [PubMed]
- Dixon, R.A.; Paiva, N.L. Stress-induced phenylpropanoid metabolism. Plant Cell 1995, 7, 1085–1097. [Google Scholar] [CrossRef] [PubMed]
- Ferrer, J.-L.; Austin, M.B.; Stewart, C., Jr.; Noel, J.P. Structure and function of enzymes involved in the biosynthesis of phenylpropanoids. Plant Physiol. Biochem. 2008, 46, 356–370. [Google Scholar] [CrossRef] [PubMed]
- Skórzyńska-Polit, E.; Drażkiewicz, M.; Krupa, Z. The Activity of the Antioxidative System in Cadmium-Treated Arabidopsis thaliana. Biol. Plant. 2003, 47, 71–78. [Google Scholar] [CrossRef]
- Ascencio-Ibáñez, J.T.; Sozzani, R.; Lee, T.J.; Chu, T.M.; Wolfinger, R.D.; Cella, R.; Hanley-Bowdoin, L. Global analysis of Arabidopsis gene expression uncovers a complex array of changes impacting pathogen response and cell cycle during geminivirus infection. Plant Physiol. 2008, 148, 436–454. [Google Scholar] [CrossRef] [PubMed]
- Gamborg, O.L.; Miller, R.A.; Ojima, K. Nutrient requirements of suspension cultures of soybean root cells. Exp. Cell Res. 1968, 50, 151–158. [Google Scholar] [CrossRef]
- Marondedze, C.; Wong, A.; Groen, A.; Serrano, N.; Jankovic, B.; Lilley, K.; Gehring, C.; Thomas, L. Exploring the Arabidopsis proteome: Influence of protein solubilization buffers on proteome coverage. Int. J. Mol. Sci. 2015, 16, 857–870. [Google Scholar] [CrossRef] [PubMed]
- Al-Shahrour, F.; Díaz-Uriarte, R.; Dopazo, J. FatiGO: A web tool for finding significant associations of Gene Ontology terms with groups of genes. Bioinformatics 2004, 20, 578–580. [Google Scholar] [CrossRef] [PubMed]
- BabelomicsV5. Available online: http://babelomics.bioinfo.cipf.es/ (accessed on 1 September 2015).


| Accession Number | Protein Name | Fold Change (Log2) | p-Value | GO Term |
|---|---|---|---|---|
| Proteins identified changing after 5 min of cAMP treatment | ||||
| At3g16460 | Jacalin-related lectin 34 | 4.495 | 0.01358 | A |
| At2g01140 | Fructose-bisphosphate aldolase 3 | 3.532 | 0.01473 | B, D, E |
| At1g20450 | Early response to dehydration 10 (ERD10) | 0.971 | 0.04245 | A, B, E |
| At2g37220 | RNA-binding protein | −0.607 | 0.00180 | A, C |
| At1g08110 | Glyoxalase I | −0.651 | 0.00912 | B, D, E |
| At1g14980 | Chaperonin 10 | −0.733 | 0.02203 | |
| At2g27710 | 60S acidic ribosomal protein family | −1.476 | 0.03411 | A |
| Proteins identified changing after 10 min of cAMP treatment | ||||
| At1g23100 | GroES-like family protein | 39.566 | 0.02049 | |
| At1g24360 | 3-Oxoacyl-[acyl-carrier-protein] reductase | 9.238 | 0.01110 | A |
| At1g28200 | FH interacting protein 1 | 2.979 | 0.04999 | |
| At1g14980 | Chaperonin 10 | 1.771 | 0.04747 | |
| At1g48920 | Nucleolin like 1 | 1.434 | 0.03283 | |
| At1g53240 | Mitochondrial malate dehydrogenase | 0.846 | 0.03931 | A, B, C, D, E |
| At2g38540 | Lipid transfer protein 1 | 0.844 | 0.04096 | |
| At2g41430 | Early response to dehydration 15 (ERD15) | 0.756 | 0.03081 | B, C, D |
| At2g47730 | Glutathione S-transferase Φ 8 | 0.726 | 0.04412 | A, B, C |
| At3g16450 | Jacalin-related lectin 33 | 0.628 | 0.02041 | A |
| At1g11580 | Methylesterase PCR A | −0.606 | 0.03673 | C |
| At4g21860 | Methionine sulfoxide reductase B2 | −0.697 | 0.04911 | |
| At4g38740 | Rotamase cyclophilin 1 (ROC1) | −0.706 | 0.00733 | D |
| At5g47200 | RAB GTPase homolog 1A | −0.856 | 0.03223 | |
| Accession Number | Protein Name | cAMP Treatment | Pathogen Response * | ||
|---|---|---|---|---|---|
| Fold Change (Log2) | p-Value | Fold Change (Log2) | p-Value | ||
| AT3G16460 | Jacalin-related lectin 34 | 4.495 | 0.01358 | 0.274 | 4.90 × 10−5 |
| AT2G01140 | Fructose-bisphosphate aldolase 3 | 3.532 | 0.01473 | 0.372 | 1.48 × 10−5 |
| AT1G20450 | Early response to dehydration 10 (ERD10) | 0.971 | 0.04245 | −0.756 | 7.00 × 10−6 |
| AT2G37220 | RNA-binding protein | −0.607 | 0.00180 | −0.539 | 2.35 × 10−5 |
| AT2G27710 | 60S acidic ribosomal protein family | −1.476 | 0.03411 | −0.692 | 2.25 × 10−7 |
| AT1G24360 | 3-Oxoacyl-[acyl-carrier-protein] reductase | 9.238 | 0.01110 | −0.265 | 3.97 × 10−4 |
| AT1G53240 | Mitochondrial malate dehydrogenase | 0.846 | 0.03931 | −0.599 | 1.11 × 10−5 |
| AT2G38540 | Lipid transfer protein 1 | 0.844 | 0.04096 | −2.701 | 5.04 × 10−25 |
| AT1G11580 | Methylesterase PCR A | −0.606 | 0.03673 | −0.194 | 1.86 × 10−3 |
| AT4G21860 | Methionine sulfoxide reductase B2 | −0.697 | 0.04911 | −0.766 | 2.55 × 10−8 |
| AT4G38740 | Rotamase cyclophilin 1 (ROC1) | −0.706 | 0.00733 | −0.827 | 1.40 × 10−7 |
| AT5G47200 | RAB GTPase homolog 1A | −0.856 | 0.03223 | 0.498 | 2.11 × 10−7 |
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Alqurashi, M.; Gehring, C.; Marondedze, C. Changes in the Arabidopsis thaliana Proteome Implicate cAMP in Biotic and Abiotic Stress Responses and Changes in Energy Metabolism. Int. J. Mol. Sci. 2016, 17, 852. https://doi.org/10.3390/ijms17060852
Alqurashi M, Gehring C, Marondedze C. Changes in the Arabidopsis thaliana Proteome Implicate cAMP in Biotic and Abiotic Stress Responses and Changes in Energy Metabolism. International Journal of Molecular Sciences. 2016; 17(6):852. https://doi.org/10.3390/ijms17060852
Chicago/Turabian StyleAlqurashi, May, Chris Gehring, and Claudius Marondedze. 2016. "Changes in the Arabidopsis thaliana Proteome Implicate cAMP in Biotic and Abiotic Stress Responses and Changes in Energy Metabolism" International Journal of Molecular Sciences 17, no. 6: 852. https://doi.org/10.3390/ijms17060852
APA StyleAlqurashi, M., Gehring, C., & Marondedze, C. (2016). Changes in the Arabidopsis thaliana Proteome Implicate cAMP in Biotic and Abiotic Stress Responses and Changes in Energy Metabolism. International Journal of Molecular Sciences, 17(6), 852. https://doi.org/10.3390/ijms17060852