Drought-Induced Leaf Proteome Changes in Switchgrass Seedlings
Abstract
:1. Introduction
2. Results
2.1. Drought-Induced Physiological Properties of Switchgrass
2.2. Effects of the ProteoMiner Enrichment Process on the Identification of Leaf Proteome
2.3. Identification of Quantified Proteins
2.4. Proteins in Regulation of Transcription and Translation
2.5. Cell Division and Cell Wall Modification
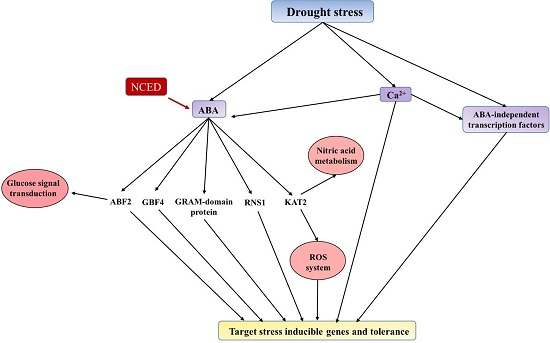
2.6. Phyto-Hormone Metabolism and Signaling Transduction Pathways
2.7. Stress-Responsive Proteins
2.8. Carbohydrate Metabolism
2.9. Nitric Acid Metabolism
3. Discussion
4. Materials and Methods
4.1. Construction of a “Sandwich” Drought Treatment System
4.2. Preparation of Seedling Plants
4.3. Drought Treatment and Physiological Measurements
4.4. Tissue Harvest and Preparation of Protein Samples
4.5. Isobaric Tags for Relative and Absolute Quantification (iTRAQ) Labeling and Mass Spectrometry Analysis
4.6. Protein Identification and Quantification, and Statistics Analysis
4.7. Functional Pathway Analysis of Drought-Induced Proteins
4.8. Statistical Analysis
5. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Wright, L.I.; Cushman, J.H.; Ehrenshaft, A.R.; McLaughlin, S.B.; McNabb, W.A.; Martin, S.A.; Ranney, J.W.; Tuskan, A.G.; Turhollow, A.F. Biofuels Feedstock Development Program Annual Progress Report for 1992; ORNL-6781; Environmental Sciences Division Publication: Washington, DC, USA, 1993. [Google Scholar]
- Parrisha, D.J.; Fike, J.H. The biology and agronomy of switchgrass for biofuels. CRC. Crit. Rev. Plant Sci. 2005, 24, 423–459. [Google Scholar] [CrossRef]
- Wright, L.L. Historical Perspective on How and Why Switchgrass Was Selected as a “Model” High-Potential Energy Crop; ORNL/TM-2007/109; Oak Ridge National Laboratory: Oak Ridge, TN, USA, 2007. [Google Scholar]
- Nezhadahmadi, A.; Prodhan, Z.H.; Faruq, G. Drought tolerance in wheat. Sci. World J. 2013, 2013, 610721. [Google Scholar] [CrossRef] [PubMed]
- National Weather Service Centers for Environmental Prediction. Available online: http://www.cpc.ncep.noaa.gov/products/monitoring_and_data/topsoil.shtml (accessed on 1 January 2016).
- Keyser, P.; Harper, C.; Bates, G.; Waller, J.; Doxon, E. Native warm-season grasses for mid-south forage production. UT Ext. 2011, SP731-A. [Google Scholar]
- Sanderson, M.A.; Reed, R.L. Switchgrass growth and development: Water, nitrogen, and plant density effects. J. Range Manag. 2000, 53, 221–227. [Google Scholar] [CrossRef]
- Vogel, K.P. Improved plant & production practices for grasslands & biomass crops in the mid-continental USA. In Proceedings of the DOE/USDA Biomass Feedstock Gate Review Meeting, Washington, DC, USA, 14–16 March 2005.
- Farooq, M.; Wahid, A.; Kobayashi, N.; Fujita, D.; Basra, S.M.A. Plant drought stress: Effects, mechanisms and management. Agron. Sustain. Dev. 2009, 29, 185–212. [Google Scholar] [CrossRef]
- Nonami, H. Plant water relations and control of cell elongation at low water potentials. J. Plant Res. 1998, 111, 373–382. [Google Scholar] [CrossRef]
- Kaya, M.D.; Okçub, G.; Ataka, M.; Çıkılıc, Y.; Kolsarıcıa, Ö. Seed treatments to overcome salt and drought stress during germination in sunflower (Helianthus annuus L.). Eur. J. Agron. 2006, 24, 291–295. [Google Scholar] [CrossRef]
- Hussain, M.; Malik, M.A.; Farooq, M.; Ashraf, M.Y.; Cheema, M.A. Improving drought tolerance by exogenous application of glycinebetaine and salicylic acid in sunflower. J. Agron. Crop Sci. 2008, 194, 193–199. [Google Scholar] [CrossRef]
- Liu, Y.; Zhang, X.; Tran, H.; Shan, L.; Kim, J.; Childs, K.; Ervin, E.H.; Frazier, T.; Zhao, B. Assessment of drought tolerance of 49 switchgrass (Panicum virgatum) genotypes using physiological and morphological parameters. Biotechnol. Biofuels 2015, 8. [Google Scholar] [CrossRef] [PubMed]
- Alberts, B.; Johnson, A.; Lewis, J. Molecular Biology of the Cell, 4th ed.; Garland Science: New York, NY, USA, 2002. [Google Scholar]
- Barkla, B.J.; Castellanos-Cervantes, T.; de León, J.L.D.; Matros, A.; Mock, H.P.; Perez-Alfocea, F.; Salekdeh, G.H.; Witzel, K.; Zörb, C. Elucidation of salt stress defense and tolerance mechanisms of crop plants using proteomics—current achievements and perspectives. Proteomics 2013, 13, 1885–1900. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, D.; Xu, J. Abiotic stress responses in plant roots: A proteomics perspective. Front. Plant Sci. 2014, 5, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Ngara, R.; Ndimba, B.K. Understanding the complex nature of salinity and drought-stress response in cereals using proteomics technologies. Proteomics 2014, 14, 611–621. [Google Scholar] [CrossRef] [PubMed]
- Okekeogbu, I.; Ye, Z.; Sangireddy, S.; Li, H.; Bhatti, S.; Hui, D.; Zhou, S.; Howe, K.; Fish, T.; Yang, Y.; et al. Effect of aluminum treatment on proteomes of radicles of seeds derived from AL-treated tomato plants. Proteomes 2014, 2, 169–190. [Google Scholar] [CrossRef]
- Zhou, S.; Sauve, R.; Fish, T.; Thannhauser, T.W. Salt-induced and Salt-suppressed Proteins in Tomato Leaves. J. Am. Soc. Hortic. Sci. 2009, 134, 289–294. [Google Scholar]
- Zhou, S.; Sauvé, R.; Liu, Z.; Reddy, S.; Bhatti, S. Heat-induced proteome changes in tomato leaves. J. Am. Soc. Hortic. Sci. 2011, 136, 219–226. [Google Scholar]
- Zhou, S.; Palmer, M.; Zhou, J.; Bhatti, S.; Howe, K.; Fish, T.; Thannhauser, T.W. Differential root proteome expression in tomato genotypes with contrasting drought tolerance exposed to dehydration. J. Am. Soc. Hort. Sci. 2013, 138, 131–141. [Google Scholar]
- Pottiez, G.; Wiederin, J.; Fox, H.S.; Ciborowski, P. Comparison of 4-plex to 8-plex iTRAQ quantitative measurements of proteins in human plasma samples. J. Proteome Res. 2012, 11, 3774–3781. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Walker, A.K.; Strahler, J.R.; Simon, E.S.; Tomanicek-Volk, S.L.; Nelson, B.B.; Hurley, M.C.; Ernst, S.; Williams, J.; Andrews, P.C. Organellar proteomics: Analysis of pancreatic zymogen granule membranes. Mol. Cell. Proteom. 2006, 5, 306–312. [Google Scholar] [CrossRef] [PubMed]
- Yang, Q.S.; Wu, J.H.; Li, C.Y.; Wei, Y.R.; Sheng, O.; Hu, C.H.; Kuang, R.-B.; Huang, Y.-H.; Peng, X.-X.; McCardle, J.; et al. Quantitative proteomic analysis reveals that antioxidation mechanisms contribute to cold tolerance in plantain (Musa paradisiaca L.; ABB Group) seedlings. Mol. Cell. Proteom. 2012, 11, 1853–1869. [Google Scholar] [CrossRef] [PubMed]
- Lan, P.; Li, W.F.; Wen, T.N.; Shiau, J.Y.; Wu, Y.C.; Lin, W.D.; Schmidt, W. iTRAQ protein profile analysis of arabidopsis roots reveals new aspects critical for iron homeostasis. Plant Physiol. 2011, 155, 821–834. [Google Scholar] [CrossRef] [PubMed]
- Redding, A.M.; Mukhopadhyay, A.; Joyner, D.C.; Hazen, T.C.; Keasling, J.D. Study of nitrate stress in desulfovibrio vulgaris hildenborough using iTRAQ proteomics. Brief. Funct. Genom. Proteom. 2006, 5, 133–143. [Google Scholar] [CrossRef] [PubMed]
- Nveawiah-Yoho, P.; Zhou, J.; Palmer, M.; Sauve, R.; Zhou, S.; Howe, K.J.; Fish, T.; Thannhauser, T.W. Identification of proteins for salt tolerance using a comparative proteomics analysis of tomato accessions with contrasting salt tolerance. J. Am. Soc. Hort. Sci. 2013, 138, 382–394. [Google Scholar]
- Nveawiah-Yoho, P. Mechanisms for Salt Tolerance and Susceptibility in Tomato. Ph.D. Thesis, Tennessee State University, Nashville, TN, USA, 2012. [Google Scholar]
- Ali, G.M.; Komatsu, S. Proteomic analysis of rice leaf sheath during drought stress. J. Proteom. Res. 2006, 5, 396–403. [Google Scholar] [CrossRef] [PubMed]
- Ke, Y.; Han, G.; He, H.; Li, J. Differential regulation of proteins and phosphoproteins in rice under drought stress. Biochem. Biophys. Res. Commun. 2009, 379, 133–138. [Google Scholar] [CrossRef] [PubMed]
- Salekdeh, G.; Siopongco, J.; Wade, L.; Ghareyazie, B.; Bennett, J. Proteomic analysis of rice leaves during drought stress and recovery. Proteomics 2002, 2, 1131–1145. [Google Scholar] [CrossRef]
- Salekdeh, G.; Siopongco, J.; Wade, L.; Ghareyazie, B.; Bennett, J. A proteomic approach to analyzing drought- and salt-responsiveness in rice. F. Crop. Res. 2002, 76, 199–219. [Google Scholar] [CrossRef]
- Shu, L.; Ding, W.; Wu, J.; Feng, F.; Luo, L. Proteomic analysis of rice leaves shows the different regulations to osmotic stress and stress signals. J. Integr. Plant Biol. 2010, 52, 981–995. [Google Scholar] [CrossRef] [PubMed]
- Xiong, J.; Fu, B.; Xu, H.; Li, Y. Proteomic analysis of PEG-simulated drought stress-responsive proteins of rice leaves using a pyramiding rice line at the seedling stage. Bot. Stud. 2010, 51, 137–145. [Google Scholar]
- De Vienne, D.; Leonardi, A.; Damerval, C.; Zivy, M. Genetics of proteome variation for QTL characterization: Application to drought-stress responses in maize. J. Exp. Bot. 1999, 50, 303–309. [Google Scholar] [CrossRef]
- Mohammadkhani, N.; Heidari, R. Effects of drought stress on soluble proteins in two maize varieties. Turkish J. Biol. 2008, 32, 23–30. [Google Scholar]
- Riccardi, F.; Gazeau, P.; de Vienne, D.; Zivy, M. Protein changes in response to progressive water deficit in maize. Plant Physiol. 1998, 117, 1253–1263. [Google Scholar] [CrossRef] [PubMed]
- McCabe, M.S.; Garratt, L.C.; Schepers, F.; Jordi, W.J.; Stoopen, G.M.; Davelaar, E.; van Rhijn, J.H.; Power, J.B.; Davey, M.R. Effects of P(SAG12)-IPT gene expression on development and senescence in transgenic lettuce. Plant Physiol. 2001, 127, 505–516. [Google Scholar] [CrossRef] [PubMed]
- Widjaja, I.; Naumann, K.; Roth, U.; Wolf, N.; Mackey, D.; Dangl, J.L.; Scheel, D.; Lee, J. Combining subproteome enrichment and Rubisco depletion enables identification of low abundance proteins differentially regulated during plant defense. Proteomics 2009, 9, 138–147. [Google Scholar] [CrossRef] [PubMed]
- Fasoli, E.; D’Amato, A.; Kravchuk, A.V.; Boschetti, E.; Bachi, A.; Righetti, P.G. Popeye strikes again: The deep proteome of spinach leaves. J. Proteom. 2011, 74, 127–136. [Google Scholar] [CrossRef] [PubMed]
- Von Toerne, C.; Kahle, M.; Schäfer, A.; Ispiryan, R.; Blindert, M.; Hrabe De Angelis, M.; Neschen, S.; Ueffing, M.; Hauck, S.M. Apoe, Mbl2, and Psp plasma protein levels correlate with diabetic phenotype in NZO mice-An optimized rapid workflow for SRM-based quantification. J. Proteome Res. 2013, 12, 1331–1343. [Google Scholar] [CrossRef] [PubMed]
- Fonslow, B.R.; Carvalho, P.C.; Academia, K.; Freeby, S.; Xu, T.; Nakorchevsky, A.; Paulus, A.; Yates, J.R. Improvements in proteomic metrics of low abundance proteins through proteome equalization using ProteoMiner prior to MudPIT. J. Proteome Res. 2011, 10, 3690–3700. [Google Scholar] [CrossRef] [PubMed]
- Polley, H.W. Implications of atmospheric and climatic change for crop yield and water use efficiency. Crop Sci. 2002, 42, 131–140. [Google Scholar] [CrossRef] [PubMed]
- Finkelstein, R.R.; Lynch, T.J. The Arabidopsis abscisic acid response gene ABI5 encodes a basic leucine zipper transcription factor. Plant Cell 2000, 12, 599–609. [Google Scholar] [CrossRef] [PubMed]
- Rösti, J.; Barton, C.J.; Albrecht, S.; Dupree, P.; Pauly, M.; Findlay, K.; Roberts, K.; Seifert, G.J. UDP-glucose 4-epimerase isoforms UGE2 and UGE4 cooperate in providing UDP-galactose for cell wall biosynthesis and growth of Arabidopsis thaliana. Plant Cell 2007, 19, 1565–1579. [Google Scholar] [CrossRef] [PubMed]
- Uozu, S.; Tanaka-Ueguchi, M.; Kitano, H.; Hattori, K.; Matsuoka, M. Characterization of XET-related genes of rice. Plant Physiol. 2000, 122, 853–859. [Google Scholar] [CrossRef] [PubMed]
- Nambara, E.; Marion-Poll, A. Abscisic acid biosynthesis and catabolism. Annu. Rev. Plant Biol. 2005, 56, 165–185. [Google Scholar] [CrossRef] [PubMed]
- Leckie, C.P.; McAinsh, M.R.; Allen, G.J.; Sanders, D.; Hetherington, A.M. Abscisic acid-induced stomatal closure mediated by cyclic ADP-ribose. Proc. Natl. Acad. Sci. USA 1998, 95, 15837–15842. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Ries, A.; Wu, K.; Yang, A.; Crawford, N.M. The Arabidopsis prohibitin gene PHB3 functions in nitric oxide-mediated responses and in hydrogen peroxide-induced nitric oxide accumulation. Plant Cell 2010, 22, 249–259. [Google Scholar] [CrossRef] [PubMed]
- García-Mata, C.; Lamattina, L. Nitric oxide induces stomatal closure and enhances the adaptive plant responses against drought stress. Plant Physiol. 2001, 126, 1196–1204. [Google Scholar] [CrossRef] [PubMed]
- Lefebvre, V.; North, H.; Frey, A.; Sotta, B.; Seo, M.; Okamoto, M.; Nambara, E.; Marion-Poll, A. Functional analysis of Arabidopsis NCED6 and NCED9 genes indicates that ABA synthesized in the endosperm is involved in the induction of seed dormancy. Plant J. 2006, 45, 309–319. [Google Scholar] [CrossRef] [PubMed]
- Huo, H.; Dahal, P.; Kunusoth, K.; McCallum, C.M.; Bradford, K.J. Expression of 9-cis-EPOXYCAROTENOID DIOXYGENASE4 is essential for thermoinhibition of lettuce seed germination but not for seed development or stress tolerance. Plant Cell 2013, 25, 884–900. [Google Scholar] [CrossRef] [PubMed]
- Tan, B.C.; Joseph, L.M.; Deng, W.T.; Liu, L.; Li, Q.B.; Cline, K.; McCarty, D.R. Molecular characterization of the Arabidopsis 9-cis epoxycarotenoid dioxygenase gene family. Plant J. 2003, 35, 44–56. [Google Scholar] [CrossRef] [PubMed]
- Seo, M.; Kanno, Y.; Frey, A.; North, H.M.; Marion-Poll, A. Dissection of Arabidopsis NCED9 promoter regulatory regions reveals a role for ABA synthesized in embryos in the regulation of GA-dependent seed germination. Plant Sci. 2016, 246, 91–97. [Google Scholar] [CrossRef] [PubMed]
- Iuchi, S.; Kobayashi, M.; Taji, T.; Naramoto, M.; Seki, M.; Kato, T. Regulation of drought tolerance by gene manipulation of 9-cis-epoxycarotenoid dioxygenase, a key enzyme in abscisic acid biosynthesis in Arabidopsis. Plant J. 2001, 27, 325–333. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.; Kang, J.; Cho, D.-I.; Park, J.H.; Kim, S.Y. ABF2, an ABRE-binding bZIP factor, is an essential component of glucose signaling and its overexpression affects multiple stress tolerance. Plant J. 2004, 40, 75–87. [Google Scholar] [CrossRef] [PubMed]
- Hillwig, M.S.; LeBrasseur, N.D.; Green, P.J.; MacIntosh, G.C. Impact of transcriptional, ABA-dependent, and ABA-independent pathways on wounding regulation of RNS1 expression. Mol. Genet. Genom. 2008, 280, 249–261. [Google Scholar] [CrossRef] [PubMed]
- Jiang, T.; Zhang, X.F.; Wang, X.F.; Zhang, D.P. Arabidopsis 3-Ketoacyl-CoA Thiolase-2 (KAT2), an enzyme of fatty acid β-Oxidation, is involved in ABA signal transduction. Plant Cell Physiol. 2011, 52, 528–538. [Google Scholar] [CrossRef] [PubMed]
- Knight, M.R.; Campbell, A.K.; Smith, S.M.; Trewavas, A.J. Transgenic plant aequorin reports the effects of touch and cold-shock and elicitors on cytoplasmic calcium. Nature 1991, 352, 524–526. [Google Scholar] [CrossRef] [PubMed]
- Knight, M.; Read, N.; Campbell, A.; Trewavas, A. Imaging calcium dynamics in living plants using semi-synthetic recombinant aequorins. J. Cell Biol. 1993, 121, 83–90. [Google Scholar] [CrossRef] [PubMed]
- Knight, M.R.; Smith, S.M.; Trewavas, A.J. Wind-induced plant motion immediately increases cytosolic calcium. Proc. Natl. Acad. Sci. USA 1992, 89, 4967–4971. [Google Scholar] [CrossRef] [PubMed]
- Braam, J. Regulated expression of the calmodulin-related TCH genes in cultured Arabidopsis cells: Induction by calcium and heat shock. Proc. Natl. Acad. Sci. USA 1992, 89, 3213–3216. [Google Scholar] [CrossRef] [PubMed]
- Haley, A.N.N.; Russell, A.J.; Wood, N.; Allan, A.C.; Knight, M.; Campbell, A.K.; Trewavas, A.J.; Ryan, C.A.; Lamb, C.J.; Jagendorf, A.T.; et al. Effects of mechanical signaling on plant cell cytosolic calcium. Proc. Natl. Acad. Sci. USA 1995, 92, 4124–4128. [Google Scholar] [CrossRef] [PubMed]
- Klee, C.; Vanaman, T. Calmodulin. Adv. Protein Chem. 1982, 35, 213–322. [Google Scholar] [PubMed]
- Roberts, D.; Lukas, T.; Watterson, D. Structure, function, and mechanisms of action of calmodulin. CRC Crit. Rev. Plant Sci. 1986, 4, 311–339. [Google Scholar] [CrossRef]
- Cohen, P.; Klee, C. Calmodulin; Elsevier Biomedical Press: Amsterdam, The Netherlands, 1988. [Google Scholar]
- Allan, E.; Hepler, P. Calmodulin and calcium-binding proteins. In The Biochemistry of Plants; Stumpf, P., Lonn, E., Eds.; Academic Press: New York, NY, USA, 1989; pp. 455–484. [Google Scholar]
- Roberts, D.M.; Harmon, A.C. Calcium modulated proteins targets of intracellular calcium signals in higher plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1992, 43, 375–414. [Google Scholar] [CrossRef]
- Maruyama, K.; Urano, K.; Yoshiwara, K.; Morishita, Y.; Sakurai, N.; Suzuki, H.; Kojima, M.; Sakakibara, H.; Shibata, D.; Saito, K.; et al. Integrated analysis of the effects of cold and dehydration on rice metabolites, phytohormones, and gene transcripts. Plant Physiol. 2014, 164, 1759–1771. [Google Scholar] [CrossRef] [PubMed]
- Madden, T.; Bally, M.; Hope, M.; Cullis, P.; Schieren, H.; Janoff, A. Protection of large unilamellar vesicles by trehalose during dehydration: Retention of vesicle contents. Biochim. Biophys. Acta 1985, 817, 67–74. [Google Scholar] [CrossRef]
- Kaplan, F.; Guy, C.L. β-amylase induction and the protective role of maltose during temperature shock. Plant Physiol. 2004, 135, 1674–1684. [Google Scholar] [CrossRef] [PubMed]
- Basu, P.S.; Ali, M.; Chaturvedi, S.K. Osmotic adjustment increases water uptake, remobilization of assimilates and maintains photosynthesis in chickpea under drought. Indian J. Exp. Biol. 2007, 45, 261–267. [Google Scholar] [PubMed]
- Kempa, S.; Krasensky, J.; Dal Santo, S.; Kopka, J.; Jonak, C. A Central Role of Abscisic Acid in Stress-Regulated Carbohydrate Metabolism. PLoS ONE 2008, 3, e3935. [Google Scholar] [CrossRef] [PubMed]
- Castonguay, Y.; Nadeau, P. Enzymatic control of soluble carbohydrate accumulation in cold-acclimated crowns of alfalfa. Crop Sci. 1998, 38, 1183–1189. [Google Scholar] [CrossRef]
- Gilmour, S.; Sebolt, A.; Salazar, M.; Everard, J.; Thomashow, M. Overexpression of the Arabidopsis CBF3 transcriptional activator mimics multiple biochemical changes associated with cold acclimation. Plant Physiol. 2000, 124, 1854–1865. [Google Scholar] [CrossRef] [PubMed]
- Taji, T.; Ohsumi, C.; Iuchi, S.; Seki, M.; Kasuga, M.; Kobayashi, M.; Yamaguchi-Shinozaki, K.; Shinozaki, K. Important roles of drought- and cold-inducible genes for galactinol synthase in stress tolerance in Arabidopsis thaliana. Plant J. 2002, 29, 417–426. [Google Scholar] [CrossRef] [PubMed]
- Cook, D.; Fowler, S.; Fiehn, O.; Thomashow, M.F. A prominent role for the CBF cold response pathway in conFigure uring the low-temperature metabolome of Arabidopsis. Proc. Natl. Acad. Sci. USA 2004, 101, 15243–15248. [Google Scholar]
- Peters, S.; Mundree, S.; Thomson, J.; Farrant, J.; Keller, F. Protection mechanisms in the resurrection plant Xerophyta viscosa (Baker): Both sucrose and raffinose family oligosaccharides (RFOs) accumulate in leaves in response to water deficit. J. Exp. Bot. 2007, 58, 1947–1956. [Google Scholar] [CrossRef] [PubMed]
- Usadel, B.; Bläsing, O.E.; Gibon, Y.; Poree, F.; Höhne, M.; Günter, M.; Trethewey, R.; Kamlage, B.; Poorter, H.; Stitt, M. Multilevel genomic analysis of the response of transcripts, enzyme activities and metabolites in Arabidopsis rosettes to a progressive decrease of temperature in the non-freezing range. Plant Cell Environ. 2008, 31, 518–547. [Google Scholar] [CrossRef] [PubMed]
- Hincha, D.K. Effects of calcium-induced aggregation on the physical stability of liposomes containing plant glycolipids. Biochim. Biophys. Acta 2003, 1611, 180–186. [Google Scholar] [CrossRef]
- Nishizawa, A.; Yabuta, Y.; Shigeoka, S. Galactinol and raffinose constitute a novel function to protect plants from oxidative damage. Plant Physiol. 2008, 147, 1251–1263. [Google Scholar] [CrossRef] [PubMed]
- Peterbauer, T.; Richter, A. Biochemistry and physiology of raffinose family oligosaccharides and galactosyl cyclitols in seeds. Seed Sci. Res. 2001, 11, 185–197. [Google Scholar]
- Rhodes, D.; Handa, S.; Bressan, R. Metabolic changes associated with adaptation of plant cells to water stress. Plant Physiol. 1986, 82, 890–903. [Google Scholar] [CrossRef] [PubMed]
- Girousse, C.; Bournoville, R.; Bonnemain, J.L. Water deficit-induced changes in concentrations in proline and some other amino acids in the phloem sap of alfalfa. Plant Physiol. 1996, 111, 109–113. [Google Scholar] [PubMed]
- Hong, Z. Removal of feedback inhibition of delta 1-pyrroline-5-carboxylate synthetase results in increased proline accumulation and protection of plants from osmotic stress. Plant Physiol. 2000, 122, 1129–1136. [Google Scholar] [CrossRef] [PubMed]
- Joshi, V.; Jander, G. Arabidopsis methionine garmma-lyase is regulated according to isoleucine biosynthesis needs but Plays a subordinate. Plant Physiol. 2009, 151, 367–378. [Google Scholar] [CrossRef] [PubMed]
- Bischoff, F.R.; Ponstingl, H. Catalysis of guanine nucleotide exchange on Ran by the mitotic regulator RCC1. Nature 1991, 354, 80–82. [Google Scholar] [CrossRef] [PubMed]
- Nemergut, M.E.; Lindsay, M.E.; Brownawell, A.M.; Macara, I.G. Ran-binding protein 3 links Crm1 to the Ran guanine nucleotide exchange factor. J. Biol. Chem. 2002, 277, 17385–17388. [Google Scholar] [CrossRef] [PubMed]
- Cho, S.K.; Kim, J.E.; Park, J.A.; Eom, T.J.; Kim, W.T. Constitutive expression of abiotic stress-inducible hot pepper CaXTH3, which encodes a xyloglucan endotransglucosylase/hydrolase homolog, improves drought and salt tolerance in transgenic Arabidopsis plants. FEBS Lett. 2006, 580, 3136–3144. [Google Scholar] [CrossRef] [PubMed]
- Thannhauser, T.; Shen, M.; Sherwood, R.; Howe, K.; Fish, T.; Yang, Y.; Chen, W.; Zhang, S. A workflow for large-scale empirical identification of cell wall N-linked glycoproteins of tomato (Solanum lycopersicum). Electrophoresis 2013, 34, 2417–2431. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Qiang, X.; Owsiany, K.; Zhang, S.; Thannhauser, T.W.; Li, L. Evaluation of different multidimensional LC-MS/MS pipelines for iTRAQ-based proteomic analysis of potato tubers in response to cold storage. J. Proteome Res. 2011, 10, 4647–4660. [Google Scholar] [CrossRef] [PubMed]
- Vizcaíno, J.A.; Csordas, A.; del-Toro, N.; Dianes, J.A.; Griss, J.; Lavidas, I.; Mayer, G.; Perez-Riverol, Y.; Reisinger, F.; Ternent, T.; et al. 2016 update of the PRIDE database and related tools. Nucleic Acids Res. 2016, 44, D447–D456. [Google Scholar] [CrossRef] [PubMed]
- Zhou, S.; Okekeogbu, I.; Sangireddy, S.; Ye, Z.; Li, H.; Bhatti, S.; Hui, D.; Mcdonald, D.W.; Yang, Y.; Giri, S.; et al. Proteome modification in tomato plants upon long-term aluminum treatment. J. Proteome Res. 2016, 15, 1670–1684. [Google Scholar] [CrossRef] [PubMed]
- MapMan, Version 3.5.1R2 2015. Available online: http://mapman.gabipd.org (accessed on 10 April 2016).


| Treatment | Control | Drought | |
|---|---|---|---|
| Soil Water Tension (MPa) | 0.00 ± 0.00 A,† | 0.08 ± 0.02 B,† | |
| Leaf Relative Water Content | 77.35 ± 0.01 A | 71.08 ± 0.02 B | |
| Plant Height (cm) | 0 Day drought treatment | 18.31 ± 6.18 A | 19.08 ± 4.97 A |
| 20 days drought treatment | 43.26 ± 9.11 A | 39.75 ± 8.49 B | |
| Relative plant height | 24.96 ± 6.21 A | 20.67 ± 6.22 B | |
| Photosynthesis | Leaf photosynthetic rate (μmol CO2/m2/s) | 22.96 ± 3.22 A | 21.69 ± 7.17 A |
| Stomatal conductance (mol H2O/m2/s) | 0.138 ± 0.03 A | 0.125 ± 0.05 B | |
| Transpiration rate (mmol H2O/m−2/s) | 6.88 ± 1.11 A | 6.09 ± 2.15 B | |
| Water use efficiency (μmol CO2/mmol H2O) | 3.35 ± 0.20 A | 3.59 ± 0.25 B | |
| Protein Classification | CLE a | PMT b | The Number of Proteins from CLE and PMT | |
|---|---|---|---|---|
| Proteins identified with one or more peptides | The total number of proteins | 5493 | 4839 | 7006 |
| The number of proteins overlapped in CLE and PMT | 3326 | |||
| The number of proteins identified in CLE | 2167 | - | ||
| The number of protein identified in PMT | - | 1513 | ||
| Quantified proteins with two or more peptides | The total number of proteins | 4746 | 4134 | 5680 |
| The number of proteins overlapped in CLE and PMT | 3200 | |||
| The number of protein in CLE | 1546 | - | ||
| The number of proteins in PMT | - | 934 | ||
| Differentially expressed proteins (FDR < 0.01, fold change < 0.06 or > 1.7) | The total number of proteins | 205 | 107 | 257 |
| The number of proteins in CLE and PMT | 55 | |||
| The number of proteins in CLE | 150 | - | ||
| The number of proteins in PMT | - | 52 | ||
| Classification | CLE a | PMT b | CLE and PMT c | |
|---|---|---|---|---|
| Molecular Function | Abiotic/biotic stress | 72 | 25 | 116 |
| Cell division/cell cycle | 11 | 7 | 42 | |
| Cell organization | 26 | 11 | 47 | |
| Cell vesicle transport | 21 | 6 | 31 | |
| Development | 41 | 16 | 46 | |
| DNA repair | 4 | 2 | 7 | |
| DNA synthesis | 20 | 15 | 28 | |
| Functional enzyme | 62 | 64 | 180 | |
| Metal binding | 4 | 1 | 11 | |
| Phyto-hormone metabolism | 21 | 11 | 36 | |
| Protein and amino acids activation | 15 | 13 | 35 | |
| Protein degradation | 88 | 39 | 172 | |
| Protein post-translation | 27 | 12 | 41 | |
| Protein synthesis | 61 | 54 | 209 | |
| Protein targeting | 21 | 22 | 81 | |
| Redox balance | 31 | 18 | 98 | |
| RNA transcription/processing | 113 | 74 | 212 | |
| Signaling regulation | 82 | 39 | 98 | |
| Transport | 24 | 35 | 65 | |
| Cellular Metabolism | Amino acid metabolism | 39 | 38 | 91 |
| C1-metabolism | 4 | 5 | 16 | |
| Cell wall synthesis/modification | 13 | 13 | 20 | |
| Fermentation | 3 | 3 | 6 | |
| Glycolysis | 9 | 12 | 41 | |
| Glyoxylate cycle | 1 | 0 | 10 | |
| Lipid metabolism | 22 | 30 | 51 | |
| Major CHO metabolism | 11 | 10 | 35 | |
| Minor CHO metabolism | 7 | 0 | 26 | |
| Mitochondrial electron transport/ATP synthesis | 9 | 9 | 57 | |
| N-metabolism | 2 | 2 | 7 | |
| Nucleotide metabolism | 24 | 14 | 53 | |
| Oxidative pentose phosphate (OPP) pathway | 7 | 3 | 12 | |
| Photosystem. Calvin cycle | 4 | 6 | 36 | |
| Photosystem. Light reaction | 15 | 10 | 82 | |
| Photorespiration | 3 | 1 | 14 | |
| S-assimilation | 2 | 2 | 5 | |
| Secondary metabolism | 18 | 29 | 67 | |
| TCA cycle | 8 | 10 | 52 | |
| Tetrapyrrole synthesis | 13 | 9 | 20 | |
| Others and not assigned proteins | 588 | 264 | 944 | |
| Total | 1546 | 934 | 3200 | |
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ye, Z.; Sangireddy, S.; Okekeogbu, I.; Zhou, S.; Yu, C.-L.; Hui, D.; Howe, K.J.; Fish, T.; Thannhauser, T.W. Drought-Induced Leaf Proteome Changes in Switchgrass Seedlings. Int. J. Mol. Sci. 2016, 17, 1251. https://doi.org/10.3390/ijms17081251
Ye Z, Sangireddy S, Okekeogbu I, Zhou S, Yu C-L, Hui D, Howe KJ, Fish T, Thannhauser TW. Drought-Induced Leaf Proteome Changes in Switchgrass Seedlings. International Journal of Molecular Sciences. 2016; 17(8):1251. https://doi.org/10.3390/ijms17081251
Chicago/Turabian StyleYe, Zhujia, Sasikiran Sangireddy, Ikenna Okekeogbu, Suping Zhou, Chih-Li Yu, Dafeng Hui, Kevin J. Howe, Tara Fish, and Theodore W. Thannhauser. 2016. "Drought-Induced Leaf Proteome Changes in Switchgrass Seedlings" International Journal of Molecular Sciences 17, no. 8: 1251. https://doi.org/10.3390/ijms17081251
APA StyleYe, Z., Sangireddy, S., Okekeogbu, I., Zhou, S., Yu, C. -L., Hui, D., Howe, K. J., Fish, T., & Thannhauser, T. W. (2016). Drought-Induced Leaf Proteome Changes in Switchgrass Seedlings. International Journal of Molecular Sciences, 17(8), 1251. https://doi.org/10.3390/ijms17081251