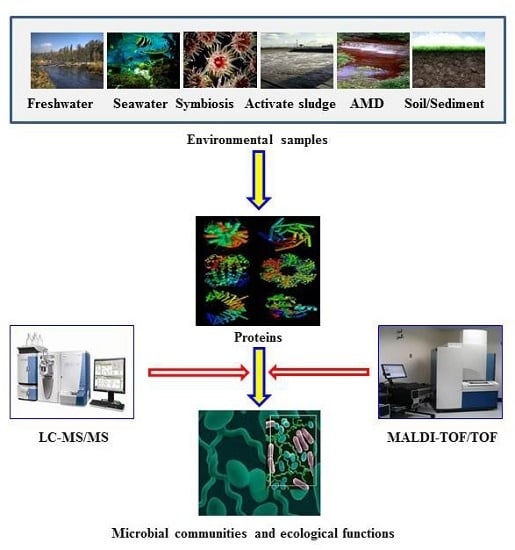
Environmental Microbial Community Proteomics: Status, Challenges and Perspectives
Abstract
:1. Introduction
2. Strategies for Microbial Community Proteomic Studies
3. Microbial Community Proteomics in Various Environments
3.1. Marine and Freshwater Metaproteomics
3.2. Soil Metaproteomics
3.3. Wastewater and Activated Sludge Metaproteomics
3.4. Acid Mine Drainage (AMD) Biofilm Metaproteomics
4. Challenges
5. Perspectives
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Williams, M.A.; Taylor, E.B.; Mula, H.P. Metaproteomic characterization of a soil microbial community following carbon amendment. Soil Biol. Biochem. 2010, 42, 1148–1156. [Google Scholar] [CrossRef]
- Wilkins, M.J.; Wrighton, K.C.; Nicora, C.D.; Williams, K.H.; McCue, L.A.; Handley, K.M.; Miller, C.S.; Giloteaux, L.; Montgomery, A.P.; Lovley, D.R.; et al. Fluctuations in species-level protein expression occur during element and nutrient cycling in the subsurface. PLoS ONE 2013, 8, e57819. [Google Scholar] [CrossRef] [PubMed]
- Segata, N.; Boernigen, D.; Tickle, T.L.; Morgan, X.C.; Garrett, W.S.; Huttenhower, C. Computational meta’omics for microbial community studies. Mol. Syst. Biol. 2013, 9, 666. [Google Scholar] [CrossRef] [PubMed]
- Wilmes, P.; Bond, P.L. The application of two-dimensional polyacrylamide gel electrophoresis and downstream analyses to a mixed community of prokaryotic microorganisms. Environ. Microbiol. 2004, 6, 911–920. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.-Z.; Xie, Z.-X.; Zhang, S.-F. Marine metaproteomics: Current status and future directions. J. Proteom. 2014, 97, 27–35. [Google Scholar] [CrossRef] [PubMed]
- Baldrian, P.; Lopez-Mondejar, R. Microbial genomics, transcriptomics and proteomics: New discoveries in decomposition research using complementary methods. Appl. Microbiol. Biotechnol. 2014, 98, 1531–1537. [Google Scholar] [CrossRef] [PubMed]
- Dyhrman, S.T. Molecular approaches to diagnosing nutritional physiology in harmful algae: Implications for studying the effects of eutrophication. Harmful Algae 2008, 8, 167–174. [Google Scholar] [CrossRef]
- Glass, J.B.; Yu, H.; Steele, J.A.; Dawson, K.S.; Sun, S.; Chourey, K.; Pan, C.; Hettich, R.L.; Orphan, V.J. Geochemical, metagenomic and metaproteomic insights into trace metal utilization by methane-oxidizing microbial consortia in sulphidic marine sediments. Environ. Microbiol. 2014, 16, 1592–1611. [Google Scholar] [CrossRef] [PubMed]
- Kleiner, M.; Young, J.C.; Shah, M.; VerBerkmoes, N.C.; Dubilier, N. Metaproteomics reveals abundant transposase expression in mutualistic endosymbionts. mBio 2013, 4, e00223–e00213. [Google Scholar] [CrossRef] [PubMed]
- Moore, C.M. Microbial proteins and oceanic nutrient cycles. Science 2014, 345, 1120–1121. [Google Scholar] [CrossRef] [PubMed]
- Schneider, T.; Riedel, K. Environmental proteomics: Analysis of structure and function of microbial communities. Proteomics 2010, 10, 785–798. [Google Scholar] [CrossRef] [PubMed]
- Schneider, T.; Gerrits, B.; Gassmann, R.; Schmid, E.; Gessner, M.O.; Richter, A.; Battin, T.; Eberl, L.; Riedel, K. Proteome analysis of fungal and bacterial involvement in leaf litter decomposition. Proteomics 2010, 10, 1819–1830. [Google Scholar] [CrossRef] [PubMed]
- Ge, Y.; Wang, D.-Z.; Chiu, J.-F.; Cristobal, S.; Sheehan, D.; Silvestre, F.; Peng, X.; Li, H.; Gong, Z.; Lam, S.H. Environmental omics: Current status and future directions. J. Integr. OMICS 2013, 3, 75–87. [Google Scholar] [CrossRef]
- Kan, J.; Hanson, T.E.; Ginter, J.M.; Wang, K.; Chen, F. Metaproteomic analysis of chesapeake bay microbial communities. Saline Syst. 2005, 1, 7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Georges, A.A.; El-Swais, H.; Craig, S.E.; Li, W.K.; Walsh, D.A. Metaproteomic analysis of a winter to spring succession in coastal northwest Atlantic Ocean microbial plankton. ISME J. 2014, 8, 1301–1313. [Google Scholar] [CrossRef] [PubMed]
- Kleiner, M.; Wentrup, C.; Lott, C.; Teeling, H.; Wetzel, S.; Young, J.; Chang, Y.J.; Shah, M.; VerBerkmoes, N.C.; Zarzycki, J.; et al. Metaproteomics of a gutless marine worm and its symbiotic microbial community reveal unusual pathways for carbon and energy use. Proc. Natl. Acad. Sci. USA 2012, 109, E1173–E1182. [Google Scholar] [CrossRef] [PubMed]
- McCarren, J.; Becker, J.W.; Repeta, D.J.; Shi, Y.; Young, C.R.; Malmstrom, R.R.; Chisholm, S.W.; DeLong, E.F. Microbial community transcriptomes reveal microbes and metabolic pathways associated with dissolved organic matter turnover in the sea. Proc. Natl. Acad. Sci. USA 2010, 107, 16420–16427. [Google Scholar] [CrossRef] [PubMed]
- Bastida, F.; Hernandez, T.; Garcia, C. Metaproteomics of soils from semiarid environment: Functional and phylogenetic information obtained with different protein extraction methods. J. Proteom. 2014, 101, 31–42. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.B.; Zhang, Z.X.; Li, H.; He, H.B.; Fang, C.X.; Zhang, A.J.; Li, Q.S.; Chen, R.S.; Guo, X.K.; Lin, H.F.; et al. Characterization of metaproteomics in crop rhizospheric soil. J. Proteome Res. 2011, 10, 932–940. [Google Scholar] [CrossRef] [PubMed]
- Martin, H.G.; Ivanova, N.; Kunin, V.; Warnecke, F.; Barry, K.W.; McHardy, A.C.; Yeates, C.; He, S.; Salamov, A.A.; Szeto, E.; et al. Metagenomic analysis of two enhanced biological phosphorus removal (EBPR) sludge communities. Nat. Biotechnol. 2006, 24, 1263–1269. [Google Scholar] [CrossRef] [PubMed]
- Wilmes, P.; Andersson, A.F.; Lefsrud, M.G.; Wexler, M.; Shah, M.; Zhang, B.; Hettich, R.L.; Bond, P.L.; VerBerkmoes, N.C.; Banfield, J.F. Community proteogenomics highlights microbial strain-variant protein expression within activated sludge performing enhanced biological phosphorus removal. ISME J. 2008, 2, 853–864. [Google Scholar] [CrossRef] [PubMed]
- Wilmes, P.; Wexler, M.; Bond, P.L. Metaproteomics provides functional insight into activated sludge wastewater treatment. PLoS ONE 2008, 3, e1778. [Google Scholar] [CrossRef] [PubMed]
- Benndorf, D.; Balcke, G.U.; Harms, H.; von Bergen, M. Functional metaproteome analysis of protein extracts from contaminated soil and groundwater. ISME J. 2007, 1, 224–234. [Google Scholar] [CrossRef] [PubMed]
- Ram, R.J.; VerBerkmoes, N.C.; Thelen, M.P.; Tyson, G.W.; Baker, B.J.; Blake, R.C.; Shah, M.; Hettich, R.L.; Banfield, J.F. Community proteomics of a natural microbial biofilm. Science 2005, 308, 1915–1920. [Google Scholar] [CrossRef] [PubMed]
- Lo, I.; Denef, V.J.; Verberkmoes, N.C.; Shah, M.B.; Goltsman, D.; DiBartolo, G.; Tyson, G.W.; Allen, E.E.; Ram, R.J.; Detter, J.C.; et al. Strain-resolved community proteomics reveals recombining genomes of acidophilic bacteria. Nature 2007, 446, 537–541. [Google Scholar] [CrossRef] [PubMed]
- Williams, T.J.; Cavicchioli, R. Marine metaproteomics: Deciphering the microbial metabolic food web. Trends Microbiol. 2014, 22, 248–260. [Google Scholar] [CrossRef] [PubMed]
- Wilmes, P.; Bond, P.L. Metaproteomics: Studying functional gene expression in microbial ecosystems. Trends Microbiol. 2006, 14, 92–97. [Google Scholar] [CrossRef] [PubMed]
- Mazard, S.; Schäfer, H. Stable isotope probing to study functional components of complex microbial ecosystems. In Environmental Microbiology; Humana Press: New York, NY, USA, 2014; pp. 169–180. [Google Scholar]
- Amann, R.; Fuchs, B.M. Single-cell identification in microbial communities by improved fluorescence in situ hybridization techniques. Nat. Rev. Microbiol. 2008, 6, 339–348. [Google Scholar] [CrossRef] [PubMed]
- Ginige, M.P.; Hugenholtz, P.; Daims, H.; Wagner, M.; Keller, J.; Blackall, L.L. Use of stable-isotope probing, full-cycle rRNA analysis, and fluorescence in situ hybridization-microautoradiography to study a methanol-fed denitrifying microbial community. Appl. Environ. Microbiol. 2004, 70, 588–596. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Rillig, M.C.; Wang, W. Improving soil protein extraction for metaproteome analysis and glomalin-related soil protein detection. Proteomics 2009, 9, 4970–4973. [Google Scholar] [CrossRef] [PubMed]
- Dong, H.-P.; Wang, D.-Z.; Dai, M.; Hong, H.-S. Characterization of particulate organic matter in the water column of the South China Sea using a shotgun proteomic approach. Limnol. Oceanogr. 2010, 55, 1565–1578. [Google Scholar] [CrossRef] [Green Version]
- Habicht, K.S.; Miller, M.; Cox, R.P.; Frigaard, N.U.; Tonolla, M.; Peduzzi, S.; Falkenby, L.G.; Andersen, J.S. Comparative proteomics and activity of a green sulfur bacterium through the water column of lake Cadagno, Switzerland. Environ. Microbiol. 2011, 13, 203–215. [Google Scholar] [CrossRef] [PubMed]
- VerBerkmoes, N.C.; Denef, V.J.; Hettich, R.L.; Banfield, J.F. Systems biology: Functional analysis of natural microbial consortia using community proteomics. Nat. Rev. Microbiol. 2009, 7, 196–205. [Google Scholar] [CrossRef] [PubMed]
- Wu, C.C.; Yates, J.R. The application of mass spectrometry to membrane proteomics. Nat. Biotechnol. 2003, 21, 262–267. [Google Scholar] [CrossRef] [PubMed]
- Elias, J.E.; Haas, W.; Faherty, B.K.; Gygi, S.P. Comparative evaluation of mass spectrometry platforms used in large-scale proteomics investigations. Nat. Methods 2005, 2, 667–675. [Google Scholar] [CrossRef] [PubMed]
- Lau, H.T.; Suh, H.W.; Golkowski, M.; Ong, S.E. Comparing silac- and stable isotope dimethyl-labeling approaches for quantitative proteomics. J. Proteome Res. 2014, 13, 4164–4174. [Google Scholar] [CrossRef] [PubMed]
- Ward, M.A.; Hyman, B.; Scherzer, C.; Trisini-Lipsanopoulos, A.; Ivinson, A.J.; Lößner, C.; Farztdinov, V.; Lovestone, S.; Pike, I. Exploration of plasma biomarkers for alzheimer’s disease using isotopic tandem mass tags and a combined directed/data-dependent acquisition NLC-MS/MS method. Alzheimer’s Dement. J. Alzheimer’s Assoc. 2014, 10, P674. [Google Scholar] [CrossRef]
- Geiger, T.; Wisniewski, J.R.; Cox, J.; Zanivan, S.; Kruger, M.; Ishihama, Y.; Mann, M. Use of stable isotope labeling by amino acids in cell culture as a spike-in standard in quantitative proteomics. Nat. Protoc. 2011, 6, 147–157. [Google Scholar] [CrossRef] [PubMed]
- Bettmer, J. Application of isotope dilution ICP-MS techniques to quantitative proteomics. Anal. Bioanal. Chem. 2010, 397, 3495–3502. [Google Scholar] [CrossRef] [PubMed]
- Hägglund, P.; Bunkenborg, J.; Maeda, K.; Svensson, B. Identification of thioredoxin disulfide targets using a quantitative proteomics approach based on isotope-coded affinity tags. J. Proteome Res. 2014, 7, 5270–5276. [Google Scholar]
- Martínez-Esteso, M.J.; Casado-Vela, J.; Sellés-Marchart, S.; Pedreño, M.A.; Bru-Martínez, R. Differential plant proteome analysis by isobaric tags for relative and absolute quantitation (iTRAQ). In Plant Proteomics; Humana Press: New York, NY, USA, 2014; pp. 155–169. [Google Scholar]
- Morris, R.M.; Nunn, B.L.; Frazar, C.; Goodlett, D.R.; Ting, Y.S.; Rocap, G. Comparative metaproteomics reveals ocean-scale shifts in microbial nutrient utilization and energy transduction. ISME J. 2010, 4, 673–685. [Google Scholar] [CrossRef] [PubMed]
- Hanson, B.T.; Hewson, I.; Madsen, E.L. Metaproteomic survey of six aquatic habitats: Discovering the identities of microbial populations active in biogeochemical cycling. Microb. Ecol. 2014, 67, 520–539. [Google Scholar] [CrossRef] [PubMed]
- Saito, M.A.; McIlvin, M.R.; Moran, D.M.; Goepfert, T.J.; DiTullio, G.R.; Post, A.F.; Lamborg, C.H. Multiple nutrient stresses at intersecting pacific ocean biomes detected by protein biomarkers. Science 2014, 345, 1173–1177. [Google Scholar] [CrossRef] [PubMed]
- Williams, T.J.; Long, E.; Evans, F.; Demaere, M.Z.; Lauro, F.M.; Raftery, M.J.; Ducklow, H.; Grzymski, J.J.; Murray, A.E.; Cavicchioli, R. A metaproteomic assessment of winter and summer bacterioplankton from Antarctic Peninsula coastal surface waters. ISME J. 2012, 6, 1883–1900. [Google Scholar] [CrossRef] [PubMed]
- Sowell, S.M.; Abraham, P.E.; Shah, M.; Verberkmoes, N.C.; Smith, D.P.; Barofsky, D.F.; Giovannoni, S.J. Environmental proteomics of microbial plankton in a highly productive coastal upwelling system. ISME J. 2011, 5, 856–865. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.-Z.; Dong, H.-P.; Xie, Z.-X.; Dai, M.-H.; Hong, H.-S. Metaproteomic characterization of dissolved organic matter in the water column of the South China Sea. Limnol. Oceanogr. 2011, 56, 1641–1652. [Google Scholar] [CrossRef]
- Grob, C.; Taubert, M.; Howat, A.M.; Burns, O.J.; Dixon, J.L.; Richnow, H.H.; Jehmlich, N.; von Bergen, M.; Chen, Y.; Murrell, J.C. Combining metagenomics with metaproteomics and stable isotope probing reveals metabolic pathways used by a naturally occurring marine methylotroph. Environ. Microbiol. 2015, 17, 4007–4018. [Google Scholar] [CrossRef] [PubMed]
- Colatriano, D.; Ramachandran, A.; Yergeau, E.; Maranger, R.; Gelinas, Y.; Walsh, D.A. Metaproteomics of aquatic microbial communities in a deep and stratified estuary. Proteomics 2015, 15, 3566–3579. [Google Scholar] [CrossRef] [PubMed]
- Ng, C.; DeMaere, M.Z.; Williams, T.J.; Lauro, F.M.; Raftery, M.; Gibson, J.A.; Andrews-Pfannkoch, C.; Lewis, M.; Hoffman, J.M.; Thomas, T.; et al. Metaproteogenomic analysis of a dominant green sulfur bacterium from Ace Lake, Antarctica. ISME J. 2010, 4, 1002–1019. [Google Scholar] [CrossRef] [PubMed]
- Lauro, F.M.; DeMaere, M.Z.; Yau, S.; Brown, M.V.; Ng, C.; Wilkins, D.; Raftery, M.J.; Gibson, J.A.; Andrews-Pfannkoch, C.; Lewis, M.; et al. An integrative study of a meromictic lake ecosystem in Antarctica. ISME J. 2011, 5, 879–895. [Google Scholar] [CrossRef] [PubMed]
- Urich, T.; Lanzen, A.; Stokke, R.; Pedersen, R.B.; Bayer, C.; Thorseth, I.H.; Schleper, C.; Steen, I.H.; Ovreas, L. Microbial community structure and functioning in marine sediments associated with diffuse hydrothermal venting assessed by integrated meta-omics. Environ. Microbiol. 2014, 16, 2699–2710. [Google Scholar] [CrossRef] [PubMed]
- Stokke, R.; Roalkvam, I.; Lanzen, A.; Haflidason, H.; Steen, I.H. Integrated metagenomic and metaproteomic analyses of an ANME-1-dominated community in marine cold seep sediments. Environ. Microbiol. 2012, 14, 1333–1346. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.; Fan, L.; Zhong, L.; Kjelleberg, S.; Thomas, T. Metaproteogenomic analysis of a community of sponge symbionts. ISME J. 2012, 6, 1515–1525. [Google Scholar] [CrossRef] [PubMed]
- Sowell, S.M.; Wilhelm, L.J.; Norbeck, A.D.; Lipton, M.S.; Nicora, C.D.; Barofsky, D.F.; Carlson, C.A.; Smith, R.D.; Giovanonni, S.J. Transport functions dominate the SAR11 metaproteome at low-nutrient extremes in the sargasso sea. ISME J. 2009, 3, 93–105. [Google Scholar] [CrossRef] [PubMed]
- Bastida, F.; García, C.; von Bergen, M.; Moreno, J.L.; Richnow, H.H.; Jehmlich, N. Deforestation fosters bacterial diversity and the cyanobacterial community responsible for carbon fixation processes under semiarid climate: A metaproteomics study. Appl. Soil Ecol. 2015, 93, 65–67. [Google Scholar] [CrossRef]
- Schneider, T.; Keiblinger, K.M.; Schmid, E.; Sterflinger-Gleixner, K.; Ellersdorfer, G.; Roschitzki, B.; Richter, A.; Eberl, L.; Zechmeister-Boltenstern, S.; Riedel, K. Who is who in litter decomposition? Metaproteomics reveals major microbial players and their biogeochemical functions. ISME J. 2012, 6, 1749–1762. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, L.; Wang, H.; Zhang, Z.; Lin, R.; Zhang, Z.; Lin, W. Comparative metaproteomic analysis on consecutively rehmannia glutinosa-monocultured rhizosphere soil. PLoS ONE 2011, 6, e20611. [Google Scholar] [CrossRef] [PubMed]
- Wilkins, M.J.; Verberkmoes, N.C.; Williams, K.H.; Callister, S.J.; Mouser, P.J.; Elifantz, H.; N’Guessan A, L.; Thomas, B.C.; Nicora, C.D.; Shah, M.B.; et al. Proteogenomic monitoring of geobacter physiology during stimulated uranium bioremediation. Appl. Environ. Microbiol. 2009, 75, 6591–6599. [Google Scholar] [CrossRef] [PubMed]
- Bastida, F.; Nicolás, C.; Moreno, J.; Hernández, T.; Garcia, C. Tracing changes in the microbial community of a hydrocarbon-polluted soil by culture-dependent proteomics. Pedosphere 2010, 20, 479–485. [Google Scholar] [CrossRef]
- Bastida, F.; Jehmlich, N.; Lima, K.; Morris, B.E.; Richnow, H.H.; Hernandez, T.; von Bergen, M.; Garcia, C. The ecological and physiological responses of the microbial community from a semiarid soil to hydrocarbon contamination and its bioremediation using compost amendment. J. Proteom. 2016, 135, 162–169. [Google Scholar] [CrossRef] [PubMed]
- Carla, M.R.; Lacerda, L.H.C.; Kenneth, F.R. Metaproteomic analysis of a bacterial community response to cadmium exposure. J. Proteome Res. 2007, 6, 1145–1152. [Google Scholar]
- Chul Park, J.T.N. Investigating the fate of activated sludge. Water Environ. Fed. 2007, 07, 1408–1421. [Google Scholar]
- Bize, A.; Cardona, L.; Desmond-Le Quemener, E.; Battimelli, A.; Badalato, N.; Bureau, C.; Madigou, C.; Chevret, D.; Guillot, A.; Monnet, V.; et al. Shotgun metaproteomic profiling of biomimetic anaerobic digestion processes treating sewage sludge. Proteomics 2015, 15, 3532–3543. [Google Scholar] [CrossRef] [PubMed]
- Denef, V.J.; VerBerkmoes, N.C.; Shah, M.B.; Abraham, P.; Lefsrud, M.; Hettich, R.L.; Banfield, J.F. Proteomics-inferred genome typing (PIGT) demonstrates inter-population recombination as a strategy for environmental adaptation. Environ. Microbiol. 2009, 11, 313–325. [Google Scholar] [CrossRef] [PubMed]
- Goltsman, D.S.; Denef, V.J.; Singer, S.W.; VerBerkmoes, N.C.; Lefsrud, M.; Mueller, R.S.; Dick, G.J.; Sun, C.L.; Wheeler, K.E.; Zemla, A.; et al. Community genomic and proteomic analyses of chemoautotrophic iron-oxidizing “leptospirillum rubarum” (group II) and “leptospirillum ferrodiazotrophum” (group III) bacteria in acid mine drainage biofilms. Appl. Environ. Microbiol. 2009, 75, 4599–4615. [Google Scholar] [CrossRef] [PubMed]
- Azam, F.; Malfatti, F. Microbial structuring of marine ecosystems. Nat. Rev. Microbiol. 2007, 5, 782–791. [Google Scholar] [CrossRef] [PubMed]
- Morris, R.M.; Rappé, M.S.; Connon, S.A.; Vergin, K.L.; Siebold, W.A.; Carlson, C.A.; Giovannoni, S.J. SAR11 clade dominates ocean surface bacterioplankton communities. Nature 2002, 420, 806–810. [Google Scholar] [CrossRef] [PubMed]
- Taylor, E.B.; Williams, M.A. Microbial protein in soil: Influence of extraction method and c amendment on extraction and recovery. Microb. Ecol. 2010, 59, 390–399. [Google Scholar] [CrossRef] [PubMed]
- Alonso-Saez, L.; Waller, A.S.; Mende, D.R.; Bakker, K.; Farnelid, H.; Yager, P.L.; Lovejoy, C.; Tremblay, J.E.; Potvin, M.; Heinrich, F.; et al. Role for urea in nitrification by polar marine archaea. Proc. Natl. Acad. Sci. USA 2012, 109, 17989–17994. [Google Scholar] [CrossRef] [PubMed]
- Yong, S.C.; Roversi, P.; Lillington, J.; Rodriguez, F.; Krehenbrink, M.; Zeldin, O.B.; Garman, E.F.; Lea, S.M.; Berks, B.C. A complex iron-calcium cofactor catalyzing phosphotransfer chemistry. Science 2014, 345, 1170–1173. [Google Scholar] [CrossRef] [PubMed]
- Cvetkovic, A.; Menon, A.L.; Thorgersen, M.P.; Scott, J.W.; Poole, F.L., 2nd; Jenney, F.E., Jr.; Lancaster, W.A.; Praissman, J.L.; Shanmukh, S.; Vaccaro, B.J.; et al. Microbial metalloproteomes are largely uncharacterized. Nature 2010, 466, 779–782. [Google Scholar] [CrossRef] [PubMed]
- Becher, D.; Bernhardt, J.; Fuchs, S.; Riedel, K. Metaproteomics to unravel major microbial players in leaf litter and soil environments: Challenges and perspectives. Proteomics 2013, 13, 2895–2909. [Google Scholar] [CrossRef] [PubMed]
- Singleton, I.; Merrington, G.; Colvan, S.; Delahunty, J.S. The potential of soil protein-based methods to indicate metal contamination. Appl. Soil Ecol. 2003, 23, 25–32. [Google Scholar] [CrossRef]
- Chourey, K.; Nissen, S.; Vishnivetskaya, T.; Shah, M.; Pfiffner, S.; Hettich, R.L.; Loffler, F.E. Environmental proteomics reveals early microbial community responses to biostimulation at a uranium- and nitrate-contaminated site. Proteomics 2013, 13, 2921–2930. [Google Scholar] [CrossRef] [PubMed]
- Wilmes, P.; Heintz-Buschart, A.; Bond, P.L. A decade of metaproteomics: Where we stand and what the future holds. Proteomics 2015, 15, 3409–3417. [Google Scholar] [CrossRef] [PubMed]
- Puigagut, J.; Salvadó, H.; García, J. Short-term harmful effects of ammonia nitrogen on activated sludge microfauna. Water Res. 2005, 39, 4397–4404. [Google Scholar] [CrossRef] [PubMed]
- Park, C.; Helm, R.F.; Novak, J.T. Investigating the fate of activated sludge extracellular proteins in sludge digestion using sodium dodecyl sulfate polyacrylamide gel electrophoresis. Water Environ. Res. 2008, 80, 2219–2227. [Google Scholar] [CrossRef] [PubMed]
- Xie, J.; He, Z.; Liu, X.; Liu, X.; van Nostrand, J.D.; Deng, Y.; Wu, L.; Zhou, J.; Qiu, G. Geochip-based analysis of the functional gene diversity and metabolic potential of microbial communities in acid mine drainage. Appl. Environ. Microbiol. 2011, 77, 991–999. [Google Scholar] [CrossRef] [PubMed]
- Banfield, J.F.; Verberkmoes, N.C.; Hettich, R.L.; Thelen, M.P. Proteogenomic approaches for the molecular characterization of natural microbial communities. Omics: J. Integr. Biol. 2005, 9, 301–333. [Google Scholar] [CrossRef] [PubMed]
- Keiblinger, K.M.; Wilhartitz, I.C.; Schneider, T.; Roschitzki, B.; Schmid, E.; Eberl, L.; Riedel, K.; Zechmeister-Boltenstern, S. Soil metaproteomics—Comparative evaluation of protein extraction protocols. Soil Biol. Biochem. 2012, 54, 14–24. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hansen, S.H.; Stensballe, A.; Nielsen, P.H.; Herbst, F.A. Metaproteomics: Evaluation of protein extraction from activated sludge. Proteomics 2014, 14, 2535–2539. [Google Scholar] [CrossRef] [PubMed]
- Leary, D.H.; Hervey, W.J.T.; Li, R.W.; Deschamps, J.R.; Kusterbeck, A.W.; Vora, G.J. Method development for metaproteomic analyses of marine biofilms. Anal. Chem. 2012, 84, 4006–4013. [Google Scholar] [CrossRef] [PubMed]
- Dong, H.-P.; Wang, D.-Z.; Dai, M.; Chan, L.L.; Hong, H.-S. Shotgun proteomics: Tools for analysis of marine particulate proteins. Limnol. Oceanogr. Methods 2009, 7, 865–874. [Google Scholar] [CrossRef]
- Jagtap, P.; McGowan, T.; Bandhakavi, S.; Tu, Z.J.; Seymour, S.; Griffin, T.J.; Rudney, J.D. Deep metaproteomic analysis of human salivary supernatant. Proteomics 2012, 12, 992–1001. [Google Scholar] [CrossRef] [PubMed]
- Maiolica, A.; Junger, M.A.; Ezkurdia, I.; Aebersold, R. Targeted proteome investigation via selected reaction monitoring mass spectrometry. J. Proteom. 2012, 75, 3495–3513. [Google Scholar] [CrossRef] [PubMed]
- Chitsaz, H.; Yee-Greenbaum, J.L.; Tesler, G.; Lombardo, M.-J.; Dupont, C.L.; Badger, J.H.; Novotny, M.; Rusch, D.B.; Fraser, L.J.; Gormley, N.A. Efficient de novo assembly of single-cell bacterial genomes from short-read data sets. Nat. Biotechnol. 2011, 29, 915–921. [Google Scholar] [CrossRef] [PubMed]
- Thrash, J.C.; Temperton, B.; Swan, B.K.; Landry, Z.C.; Woyke, T.; DeLong, E.F.; Stepanauskas, R.; Giovannoni, S.J. Single-cell enabled comparative genomics of a deep ocean SAR11 bathytype. ISME J. 2014, 8, 1440–1451. [Google Scholar] [CrossRef] [PubMed]
- Kobir, A.; Shi, L.; Boskovic, A.; Grangeasse, C.; Franjevic, D.; Mijakovic, I. Protein phosphorylation in bacterial signal transduction. Biochim. Biophys. Acta 2011, 1810, 989–994. [Google Scholar] [CrossRef] [PubMed]

| Environment | Subject of Analysis | Protein-Method | Major Findings | Refs/Year | |
|---|---|---|---|---|---|
| Separation | Identification | ||||
| Marine and Freshwater | |||||
| South Atlantic | Microbial membrane proteins from surface water | LC-MS/MS | TonB-dependent transporters dominated bacterial membrane proteins while bacterial rhodopsins were detected in every sample; Archaeal, ammonia monooxygenase proteins were identified in upwelling region. | [43]/2010 | |
| The Northwest Atlantic Ocean | Metabolic activity of microbial plankton in a seasonally hypoxic basin | LC-MS/MS | A seasonal increase in high-affinity membrane transport proteins involved in scavenging of organic substrates; Rhodobacterales transporters were strongly associated with the spring phytoplankton bloom, whereas SAR11 transporters were abundant in the underlying waters. | [15]/2014 | |
| Six disparate aquatic habitats | Microbial populations | LC-MS/MS | ABC-type sugar-, organic polyanion–, and glycine betaine–transport proteins were identified from Pelagibacter, and these transporters play important roles in carbon and nitrogen cycling. | [44]/2014 | |
| The intersecting Pacific Ocean | Multiple nutrients | MS-based Multiple Reaction Monitoring | Nitrogen response regulator NtcA was abundant, which was consistent with the prevalence of the Prochlorococcus urea transporter proteins (UrtA) in low-nitrogen areas. | [45]/2014 | |
| The Antarctic Peninsula coast | Bacterioplankton of winter and summer | SDS–PAGE | MS/MS | Roseobacter clade contributed a large portion of ABC transporter for amino acids and polyamines; transporter proteins involved in amino acid, taurine and polyamine transport, as well as glutamine synthetase, were highly detected in SAR11; proteins involved in two chemolithoautotrophic pathways dominated the winter metaproteome of cold and dark polar water. | [46]/2012 |
| The Oregon coast | Microbial plankton of upwelling region | 2D-LC-MS/MS | Thirty-six percent and 17% of detected proteins were from the SAR11 clade and Roseobacter clade; transporters for amino acids, taurine, polyamines and glutamine synthetase were highly detected; methanol dehydrogenase was detected. | [47]/2011 | |
| Symbionts | A gutless worm and its symbiotic microbial community | 1D PAGE; 2D | LC-MS/MS | Sulfur oxidation proteins; aerobic and anaerobic CO dehydrogenases were detected in three types of Olavius algarvensis symbionts; high expression of periplasmic uptake (NiFeSe) hydrogenases and high-affinity uptake transport related proteins were detected. | [16]/2013 |
| The South China Sea | Dissolved organic matter (DOM) from marine surface and bathypelagic region | SDS-PAGE | LC-MS/MS | Archaea and Proteobacteria were the major contributors to bathypelagic proteome; protein compositions differed along the vertical water column, and urea ABC transporter was abundant in the surface DOM. | [48]/2011 |
| The English Channel | Natural populations | Protein-SIP LC-MS/MS | RuMP cycle was the main carbon assimilation pathway in Methylophaga-like bacterium, and methanol dehydrogenase–encoding gene mxaF, as well as three out of four identified xoxF homologues were expressed. | [49]/2015 | |
| The Lower St. Lawrence Estuary | Microbial communities through the stratified water column | LC-MS/MS | Chemosynthetic production coupled to nitrification by MG-I Thaumarchaeota and Nitrospina was a dominant metabolic strategy; methanol oxidation proteins were detected from the OM43 marine clade; membrane transport proteins were assigned to the uncultivated MG-II Euryarchaeota. | [50]/2015 | |
| Sulphidic marine sediments | Trace metal utilization of methane-oxidizing microbial consortia | LC-MS/MS | Microbial consortia relied on the nickel metalloenzymes and transporters, cobalt metalloenzymes and transporters, molybdenum and tungsten enzymes to catalyze anaerobic oxidation of methane (AOM). | [8]/2014 | |
| Ace Lake in Antarctica | Green sulfur bacteria | SDS-PAGE | LC-MS/MS | Proteins that participated in DNA processing, nucleic acid binding, folding/refolding of proteins and lipid biosynthesis were identified to be involved in cold adaption of green sulfur bacteria. | [51]/2010 [52]/2011 |
| The meromictic Lake Cadagno | Green sulfur Bacterium Chlorobium clathratiforme | LC-MS/MS | Chlorobium clathratiforme contained enzymes for fixation of N(2) and oxidation of sulfide to sulfate, and they were not active in the dark; fermentation of polyglucose in the dark was the major pathway to obtain energy. | [33]/2011 | |
| Hydrothermal venting sediments | Microbial community structure and functioning | SDS-PAGE | LTQ Orbitrap-MS/MS | Epsilonpro-teobacteria, δ- and γ-proteobacteria, ciliates, nematodes and various archaeal taxa were identified; high expressions of carbon fixation pathways as well as chemotaxis and flagella genes. | [53]/2014 |
| Marine seep sediments | Free-living ANME-1; Sulfate-reducing bacteria | 2-DE | MS | Anaerobic methanotrophic archaea dominated microbial species involved in the sulfur cycle and the biological sinking of methane; cold-adaptation proteins and key metabolic enzymes involved in the reverse methanogenesis and sulfate-reduction pathways were identified. | [54]/2012 |
| Symbionts | Microbial community of the sponge Cymbastela | LC-MS | Proteins involved in cold adaptation and production of gas vesicles were abundant; high expressions of affinity transporters and alternative energy–utilizing proteins under stress conditions. | [55]/2012 | |
| The Sargasso Sea | Microbial membrane proteins of surface water; the SAR11 clade | LC-MS/MS | SAR11 periplasmic substrate-binding proteins (PBP) for phosphate were most abundant; proteins involved in amino acids, phosphonate, sugars and spermidine were detected. | [56]/2009 | |
| The western South China Sea | Particulate organic matters (POM) from marine surface and mesopelagic layers | SDS-PAGE | LC-MS/MS | Cyanobacteria was the largest contributor; photosynthesis-associated proteins; porins, adenosine triphosphate synthases, nutrient transporters, molecular chaperones, and ectoenzymes were detected. | [32]/2010 |
| Soils | |||||
| Semiarid soils | Functional and phylogenetic information | SDS-PAGE | LC-MS-MS | Three protein extraction methods were examined, and the functional, phylogenetic and bio-geochemical information obtained by three methods in semiarid soils presented distinct edaphic properties. | [18]/2014 |
| Semiarid soils | Deforestation fosters bacterial diversity and the cyanobacterial community | SDS-PAGE | LC-MS-MS | Deforestation increased bacterial diversity in semiarid ecosystems and raised the abundance of cyanobacterial proteins involved in C-fixation in semiarid areas. | [57]/2015 |
| Beech leaf litter | Environmental factors and nutrients on the decomposer structure and function | SDS-PAGE | LC-MS/MS | Fungi were the main producers of extracellular hydrolytic enzymes, and microbial activity was stimulated at a higher litter nutrient content via a higher abundance and activity of extracellular enzymes. | [58]/2012 |
| Crop rhizospheric soil | Crop soil metaproteomics | 2-DE | MALDI-TOF/TOF-MS | Proteins involved in protein, energy, nucleotide, secondary metabolisms and signal transduction and resistance were identified; most upregulated plant proteins were involved in carbon and nitrogen metabolism and stress response, while the majority of the upregulated microbial proteins participated in protein metabolism and cell-wall biosynthesis. | [19]/2011 [59]/2011 |
| Toluene-amended soil | The microbial community proteome | SDS–PAGE | MALDI-MS | Glutamine synthetase (Gln), ABC transporters, extracellular solute-binding proteins, outer membrane proteins (Omp) were upregulated in toluene-amended soil; arginine deiminase (ArcA) and cold-shock protein presented in toluene-amended culture while superoxide dismutase (SodB) and chaperonin (GroEL) presented in toluene-amended soil. | [1]/2010 |
| Humic soil | Enzymes connected with bacterial metabolic pathways | SDS-PAGE; 2-DE | LC-ESI-MS | Protein extraction method from soil was developed, and 2,4-dichlorophenoxy acetate dioxygenase, chlorocatechol dioxygenases, molecular chaperons and transcription factors were identified. | [23]/2007 |
| Uranium-amended soil | The subsurface microbial communities | 2-DE | LC-MS-MS | The proteome was dominated by the enzymes converting acetate to acetyl-coenzyme A and pyruvate for central metabolism. Geobacter dominated the microbial community and they participated in energy generation during biostimulation. | [60]/2009 |
| Hydrocarbon-polluted soil | Changes in the microbial community | SDS-PAGE | HPLC-MS/MS | The complexity of the microbial community showed a relative increase in hydrocarbon-enriched cultures, and the majority of identified proteins were related to glycolysis pathways, structural or protein synthesis. | [61]/2010 |
| Hydrocarbon-polluted soil | Changes in the microbial community | SDS-PAGE | LC-MS/MS | Proteobacterial protein expressions increased while the abundance of Rhizobiales decreased during petroleum pollution; compost-assisted bioremediation was mainly driven by Sphingomonadales; abundances of catechol 2,3-dioxygenases, cis-dihydrodiol dehydrogenase and 2-hydroxymuconic semialdehyde were increased. | [62]/2016 |
| Wastewater and Activated Sludge | |||||
| Cadmium-polluted wastewater | The response of a natural community | 2-DE | MALDI-TOF/TOF MS | Significant community proteome responses to cadmium exposure were observed, and ATPases, oxidoreductases, and transport proteins played important roles in the cadmium shock. | [63]/2007 |
| Wastewater sludge | Laboratory wastewater sludge microbial communities | 2-D PAGE | MALDI-TOF-MS | Substantial differences in protein abundance for enzyme variants were uncovered among the A. phosphatis population, and these proteins were mainly involved in core metabolism, EBPR-specific pathways, energy generation. | [4]/2004 [21]/2008 [22]/2008 |
| Wastewater activated sludge | Extracellular proteins in sludge digestion | SDS-PAGE | LC-MS/MS | The proteins resistant to degradation and generated during anaerobic digestion were identified, including a limited number of bacterial and human polypeptides. | [64]/2008 |
| Sewage sludge | Different proteins in two parallel anaerobic digestion lines | SDS-PAGE | LC-MS/MS | Protein-inferred and 16S rDNA tags–based taxonomic community profiles were not consistent, and a high proportion of proteins belonged to “Candidatus Competibacter” group. | [65]/2015 |
| Acid Mine Drainage Biofilm | |||||
| Acid mine drainage biofilm(AMD) | Gene expression, identified key activities, examined partitioning of metabolic functions | 2-DE | LC-MS/MS | Half of the predicted proteins from the dominant biofilm organism Leptospirillum group II and protein involved in refolding and oxidative stress response presented high expressions; cytochrome played a central role in iron oxidation and AMD formation. | [24]/2005 |
| Acid mine drainage biofilm | Biofilms growing at the liquid-air interface | 2-DE | LC-MS/MS | Leptospirillum groups II and III dominated AMD system; signal transduction and methyl-accepting chemotaxis proteins were abundant in Leptospirillum group III, while Leptospirillum group III possessed a methyl-independent response pathway. | [66]/2009 [67]/2009 |
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, D.-Z.; Kong, L.-F.; Li, Y.-Y.; Xie, Z.-X. Environmental Microbial Community Proteomics: Status, Challenges and Perspectives. Int. J. Mol. Sci. 2016, 17, 1275. https://doi.org/10.3390/ijms17081275
Wang D-Z, Kong L-F, Li Y-Y, Xie Z-X. Environmental Microbial Community Proteomics: Status, Challenges and Perspectives. International Journal of Molecular Sciences. 2016; 17(8):1275. https://doi.org/10.3390/ijms17081275
Chicago/Turabian StyleWang, Da-Zhi, Ling-Fen Kong, Yuan-Yuan Li, and Zhang-Xian Xie. 2016. "Environmental Microbial Community Proteomics: Status, Challenges and Perspectives" International Journal of Molecular Sciences 17, no. 8: 1275. https://doi.org/10.3390/ijms17081275
APA StyleWang, D.-Z., Kong, L.-F., Li, Y.-Y., & Xie, Z.-X. (2016). Environmental Microbial Community Proteomics: Status, Challenges and Perspectives. International Journal of Molecular Sciences, 17(8), 1275. https://doi.org/10.3390/ijms17081275