Using Proteomics to Understand How Leishmania Parasites Survive inside the Host and Establish Infection
Abstract
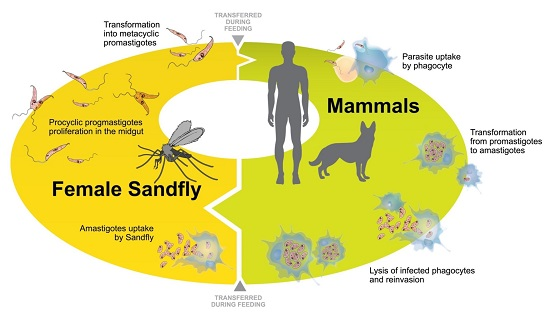
:1. Introduction
2. Leishmania Adaptation to the Intracellular Life-Cycle: Modulation in Parasite Protein Expression
2.1. Modulation of Proteins during Axenic Differentiation of Leishmania Parasites
2.2. Modulation of Proteins during Intracellular Differentiation of Leishmania Parasites
3. Protein Expression by Macrophages in Response to Leishmania Infection in Vitro
4. Protein Profiling in Human Cutaneous Lesions
5. Protein Expression in Serum of Individuals with Visceral Leishmaniasis
6. Concluding Remarks
Acknowledgments
Conflicts of Interest
References
- Alvar, J.; Velez, I.D.; Bern, C.; Herrero, M.; Desjeux, P.; Cano, J.; Jannin, J.; den Boer, M.; WHO. Leishmaniasis Control Team. Leishmaniasis worldwide and global estimates of its incidence. PLoS ONE 2012, 7, e35671. [Google Scholar] [CrossRef] [PubMed]
- WHO. Leishmaniasis. Situation and Trends. Available online: http://apps.who.int/gho/data/node.main.NTDLEISH?lang=en (accessed on 1 August 2016).
- Zijlstra, E.E.; Musa, A.M.; Khalil, E.A.; El-Hassan, I.M.; El-Hassan, A.M. Post-kala-azar dermal leishmaniasis. Lancet Infect. Dis. 2003, 3, 87–98. [Google Scholar] [CrossRef]
- Desjeux, P. Human leishmaniases: Epidemiology and public health aspects. World Health Stat. Q. 1992, 45, 267–275. [Google Scholar] [PubMed]
- Graves, P.R.; Haystead, T.A. Molecular biologist’s guide to proteomics. Microbiol. Mol. Biol. Rev. 2002, 66, 39–63. [Google Scholar] [CrossRef] [PubMed]
- Khurshid, Z.; Zohaib, S.; Najeeb, S.; Zafar, M.S.; Rehman, R.; Rehman, I.U. Advances of proteomic sciences in dentistry. Int. J. Mol. Sci. 2016, 17, 728. [Google Scholar] [CrossRef] [PubMed]
- Renella, R. Clinically-oriented proteomic investigation of sickle cell disease: Opportunities and challenges. Proteom. Clin. Appl. 2016. [Google Scholar] [CrossRef] [PubMed]
- Gaudreau, P.O.; Stagg, J.; Soulieres, D.; Saad, F. The present and future of biomarkers in prostate cancer: Proteomics, genomics, and immunology advancements. Biomark. Cancer 2016, 8, 15–33. [Google Scholar] [CrossRef] [PubMed]
- Ahsan, N.; Rao, R.S.; Gruppuso, P.A.; Ramratnam, B.; Salomon, A.R. Targeted proteomics: Current status and future perspectives for quantification of food allergens. J. Proteom. 2016, 143, 15–23. [Google Scholar] [CrossRef] [PubMed]
- Ferreira, D.; Seca, A.M.; Diana, C.G.A.; Silva, A.M. Targeting human pathogenic bacteria by siderophores: A proteomics review. J. Proteom. 2016, 16. [Google Scholar] [CrossRef] [PubMed]
- Charest, H.; Zhang, W.W.; Matlashewski, G. The developmental expression of Leishmania donovani A2 amastigote-specific genes is post-transcriptionally mediated and involves elements located in the 3′-untranslated region. J. Biol. Chem. 1996, 271, 17081–17090. [Google Scholar] [CrossRef] [PubMed]
- Brooks, D.R.; Denise, H.; Westrop, G.D.; Coombs, G.H.; Mottram, J.C. The stage-regulated expression of Leishmania mexicana CPB cysteine proteases is mediated by an intercistronic sequence element. J. Biol. Chem. 2001, 276, 47061–47069. [Google Scholar] [CrossRef] [PubMed]
- Kelly, B.L.; Nelson, T.N.; McMaster, W.R. Stage-specific expression in leishmania conferred by 3′ untranslated regions of L. Major Leishmanolysin genes (GP63). Mol. Biochem. Parasitol. 2001, 116, 101–104. [Google Scholar] [PubMed]
- Clayton, C.; Shapira, M. Post-transcriptional regulation of gene expression in trypanosomes and Leishmanias. Mol. Biochem. Parasitol. 2007, 156, 93–101. [Google Scholar] [CrossRef] [PubMed]
- Sodre, C.L.; Chapeaurouge, A.D.; Kalume, D.E.; de Mendonca Lima, L.; Perales, J.; Fernandes, O. Proteomic map of Trypanosoma cruzi CL brener: The reference strain of the genome project. Arch. Microbiol. 2009, 191, 177–184. [Google Scholar] [CrossRef] [PubMed]
- Gygi, S.P.; Rochon, Y.; Franza, B.R.; Aebersold, R. Correlation between protein and mRNA abundance in yeast. Mol. Cell. Biol. 1999, 19, 1720–1730. [Google Scholar] [CrossRef] [PubMed]
- Larreta, R.; Soto, M.; Quijada, L.; Folgueira, C.; Abanades, D.R.; Alonso, C.; Requena, J.M. The expression of HSP83 genes in Leishmania infantum is affected by temperature and by stage-differentiation and is regulated at the levels of mrna stability and translation. BMC Mol. Biol. 2004, 5, 3. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- El Fakhry, Y.; Ouellette, M.; Papadopoulou, B. A proteomic approach to identify developmentally regulated proteins in Leishmania infantum. Proteomics 2002, 2, 1007–1017. [Google Scholar] [CrossRef]
- Bente, M.; Harder, S.; Wiesgigl, M.; Heukeshoven, J.; Gelhaus, C.; Krause, E.; Clos, J.; Bruchhaus, I. Developmentally induced changes of the proteome in the protozoan parasite Leishmania donovani. Proteomics 2003, 3, 1811–1829. [Google Scholar] [CrossRef] [PubMed]
- Morales, M.A.; Watanabe, R.; Laurent, C.; Lenormand, P.; Rousselle, J.C.; Namane, A.; Spath, G.F. Phosphoproteomic analysis of Leishmania donovani pro- and amastigote stages. Proteomics 2008, 8, 350–363. [Google Scholar] [CrossRef] [PubMed]
- Biyani, N.; Madhubala, R. Quantitative proteomic profiling of the promastigotes and the intracellular amastigotes of Leishmania donovani isolates identifies novel proteins having a role in leishmania differentiation and intracellular survival. Biochim. Biophys. Acta 2012, 1824, 1342–1350. [Google Scholar] [CrossRef] [PubMed]
- Menezes, J.P.; Almeida, T.F.; Petersen, A.L.; Guedes, C.E.; Mota, M.S.; Lima, J.G.; Palma, L.C.; Buck, G.A.; Krieger, M.A.; Probst, C.M.; et al. Proteomic analysis reveals differentially expressed proteins in macrophages infected with Leishmania amazonensis or Leishmania major. Microbes Infect. 2013, 15, 579–591. [Google Scholar] [CrossRef] [PubMed]
- Singh, A.K.; Pandey, R.K.; Siqueira-Neto, J.L.; Kwon, Y.J.; Freitas-Junior, L.H.; Shaha, C.; Madhubala, R. Proteomic-based approach to gain insight into reprogramming of THP-1 cells exposed to Leishmania donovani over an early temporal window. Infect. Immun. 2015, 83, 1853–1868. [Google Scholar] [CrossRef] [PubMed]
- Costa, M.M.; Andrade, H.M.; Bartholomeu, D.C.; Freitas, L.M.; Pires, S.F.; Chapeaurouge, A.D.; Perales, J.; Ferreira, A.T.; Giusta, M.S.; Melo, M.N.; et al. Analysis of Leishmania chagasi by 2-D difference gel electrophoresis (2-D dige) and immunoproteomic: Identification of novel candidate antigens for diagnostic tests and vaccine. J. Proteome Res. 2011, 10, 2172–2184. [Google Scholar] [CrossRef] [PubMed]
- Paape, D.; Aebischer, T. Contribution of proteomics of Leishmania spp. To the understanding of differentiation, drug resistance mechanisms, vaccine and drug development. J. Proteom. 2011, 74, 1614–1624. [Google Scholar] [CrossRef] [PubMed]
- Da Silva Santos, C.; Attarha, S.; Saini, R.K.; Boaventura, V.; Costa, J.; Khouri, R.; Barral-Netto, M.; Brodskyn, C.I.; Souchelnytskyi, S. Proteome profiling of human cutaneous leishmaniasis lesion. J. Investig. Dermatol. 2015, 135, 400–410. [Google Scholar] [CrossRef] [PubMed]
- Britti, D.; Gaspari, M.; Massimini, G.; Casalinuovo, F.; Morittu, V.M.; Cuda, G. Proteomic analysis in canine leishmaniasis. Vet. Res. Commun. 2010, 34 (Suppl. 1), S91–S96. [Google Scholar] [CrossRef] [PubMed]
- Rukmangadachar, L.A.; Kataria, J.; Hariprasad, G.; Samantaray, J.C.; Srinivasan, A. Two-dimensional difference gel electrophoresis (DIGE) analysis of sera from visceral leishmaniasis patients. Clin. Proteom. 2011, 8, 4. [Google Scholar] [CrossRef] [PubMed]
- Bag, A.K.; Saha, S.; Sundar, S.; Saha, B.; Chakrabarti, A.; Mandal, C. Comparative proteomics and glycoproteomics of plasma proteins in Indian visceral leishmaniasis. Proteome Sci. 2014, 12, 48. [Google Scholar] [CrossRef] [PubMed]
- MacFarlane, J.; Blaxter, M.L.; Bishop, R.P.; Miles, M.A.; Kelly, J.M. Identification and characterisation of a Leishmania donovani antigen belonging to the 70-kDa heat-shock protein family. Eur. J. Biochem. 1990, 190, 377–384. [Google Scholar] [CrossRef] [PubMed]
- Glaser, T.A.; Moody, S.F.; Handman, E.; Bacic, A.; Spithill, T.W. An antigenically distinct lipophosphoglycan on amastigotes of Leishmania major. Mol. Biochem. Parasitol. 1991, 45, 337–344. [Google Scholar] [CrossRef]
- Zilberstein, D.; Shapira, M. The role of pH and temperature in the development of Leishmania parasites. Annu. Rev. Microbiol. 1994, 48, 449–470. [Google Scholar] [CrossRef] [PubMed]
- Alexander, J.; Russell, D.G. The interaction of leishmania species with macrophages. Adv. Parasitol. 1992, 31, 175–254. [Google Scholar] [PubMed]
- Chappuis, F.; Sundar, S.; Hailu, A.; Ghalib, H.; Rijal, S.; Peeling, R.W.; Alvar, J.; Boelaert, M. Visceral leishmaniasis: What are the needs for diagnosis, treatment and control? Nat. Rev. Microbiol. 2007, 5, 873–882. [Google Scholar] [CrossRef] [PubMed]
- Bates, P.A.; Rogers, M.E. New insights into the developmental biology and transmission mechanisms of Leishmania. Curr. Mol. Med. 2004, 4, 601–609. [Google Scholar] [CrossRef] [PubMed]
- Kaye, P.; Scott, P. Leishmaniasis: Complexity at the host-pathogen interface. Nat. Rev. Microbiol. 2011, 9, 604–615. [Google Scholar] [CrossRef] [PubMed]
- Matlashewski, G. Leishmania infection and virulence. Med. Microbiol. Immunol. 2001, 190, 37–42. [Google Scholar] [CrossRef] [PubMed]
- Turco, S.J.; Spath, G.F.; Beverley, S.M. Is lipophosphoglycan a virulence factor? A surprising diversity between Leishmania species. Trends Parasitol. 2001, 17, 223–226. [Google Scholar] [CrossRef]
- Liaud, M.F.; Lichtle, C.; Apt, K.; Martin, W.; Cerff, R. Compartment-specific isoforms of TPI and GAPDH are imported into diatom mitochondria as a fusion protein: Evidence in favor of a mitochondrial origin of the eukaryotic glycolytic pathway. Mol. Biol. Evol. 2000, 17, 213–223. [Google Scholar] [CrossRef] [PubMed]
- McNicoll, F.; Drummelsmith, J.; Muller, M.; Madore, E.; Boilard, N.; Ouellette, M.; Papadopoulou, B. A combined proteomic and transcriptomic approach to the study of stage differentiation in Leishmania infantum. Proteomics 2006, 6, 3567–3581. [Google Scholar] [CrossRef] [PubMed]
- Nugent, P.G.; Karsani, S.A.; Wait, R.; Tempero, J.; Smith, D.F. Proteomic analysis of Leishmania mexicana differentiation. Mol. Biochem. Parasitol. 2004, 136, 51–62. [Google Scholar] [CrossRef] [PubMed]
- Walker, J.; Vasquez, J.J.; Gomez, M.A.; Drummelsmith, J.; Burchmore, R.; Girard, I.; Ouellette, M. Identification of developmentally-regulated proteins in Leishmania panamensis by proteome profiling of promastigotes and axenic amastigotes. Mol. Biochem. Parasitol. 2006, 147, 64–73. [Google Scholar] [CrossRef] [PubMed]
- Brotherton, M.C.; Racine, G.; Foucher, A.L.; Drummelsmith, J.; Papadopoulou, B.; Ouellette, M. Analysis of stage-specific expression of basic proteins in Leishmania infantum. J. Proteome Res. 2010, 9, 3842–3853. [Google Scholar] [CrossRef] [PubMed]
- Acestor, N.; Masina, S.; Walker, J.; Saravia, N.G.; Fasel, N.; Quadroni, M. Establishing two-dimensional gels for the analysis of Leishmania proteomes. Proteomics 2002, 2, 877–879. [Google Scholar] [CrossRef]
- Leifso, K.; Cohen-Freue, G.; Dogra, N.; Murray, A.; McMaster, W.R. Genomic and proteomic expression analysis of Leishmania promastigote and amastigote life stages: The Leishmania genome is constitutively expressed. Mol. Biochem. Parasitol. 2007, 152, 35–46. [Google Scholar] [CrossRef] [PubMed]
- Pan, A.A.; Duboise, S.M.; Eperon, S.; Rivas, L.; Hodgkinson, V.; Traub-Cseko, Y.; McMahon-Pratt, D. Developmental life cycle of Leishmania—Cultivation and characterization of cultured extracellular amastigotes. J. Eukaryot. Microbiol. 1993, 40, 213–223. [Google Scholar] [CrossRef] [PubMed]
- Sereno, D.; Lemesre, J.L. Axenically cultured amastigote forms as an in vitro model for investigation of antileishmanial agents. Antimicrob. Agents Chemother. 1997, 41, 972–976. [Google Scholar] [PubMed]
- Puentes, F.; Diaz, D.; Hoya, R.D.; Gutierrez, J.A.; Lozano, J.M.; Patarroyo, M.E.; Moreno, A. Cultivation and characterization of stable Leishmania guyanensis complex axenic amastigotes derived from infected U937 cells. Am. J. Trop. Med. Hyg. 2000, 63, 102–110. [Google Scholar] [PubMed]
- Gupta, N.; Goyal, N.; Rastogi, A.K. In vitro cultivation and characterization of axenic amastigotes of Leishmania. Trends Parasitol. 2001, 17, 150–153. [Google Scholar] [CrossRef]
- Sereno, D.; Roy, G.; Lemesre, J.L.; Papadopoulou, B.; Ouellette, M. DNA transformation of Leishmania infantum axenic amastigotes and their use in drug screening. Antimicrob. Agents Chemother. 2001, 45, 1168–1173. [Google Scholar] [CrossRef] [PubMed]
- Debrabant, A.; Joshi, M.B.; Pimenta, P.F.; Dwyer, D.M. Generation of Leishmania donovani axenic amastigotes: Their growth and biological characteristics. Int. J. Parasitol. 2004, 34, 205–217. [Google Scholar] [CrossRef] [PubMed]
- Paape, D.; Lippuner, C.; Schmid, M.; Ackermann, R.; Barrios-Llerena, M.E.; Zimny-Arndt, U.; Brinkmann, V.; Arndt, B.; Pleissner, K.P.; Jungblut, P.R.; et al. Transgenic, fluorescent Leishmania mexicana allow direct analysis of the proteome of intracellular amastigotes. Mol. Cell. Proteom. 2008, 7, 1688–1701. [Google Scholar] [CrossRef] [PubMed]
- Rosenzweig, D.; Smith, D.; Myler, P.J.; Olafson, R.W.; Zilberstein, D. Post-translational modification of cellular proteins during Leishmania donovani differentiation. Proteomics 2008, 8, 1843–1850. [Google Scholar] [CrossRef] [PubMed]
- Purdy, J.E.; Donelson, J.E.; Wilson, M.E. Regulation of genes encoding the major surface protease of Leishmania chagasi via mRNA stability. Mol. Biochem. Parasitol. 2005, 142, 88–97. [Google Scholar] [CrossRef] [PubMed]
- Gomes, I.N.; Calabrich, A.F.; Tavares Rda, S.; Wietzerbin, J.; de Freitas, L.A.; Veras, P.S. Differential properties of CBA/J mononuclear phagocytes recovered from an inflammatory site and probed with two different species of Leishmania. Microbes Infect. 2003, 5, 251–260. [Google Scholar] [CrossRef]
- Wang, L.; Cummings, R.; Usatyuk, P.; Morris, A.; Irani, K.; Natarajan, V. Involvement of phospholipases D1 and D2 in sphingosine 1-phosphate-induced ERK (extracellular-signal-regulated kinase) activation and interleukin-8 secretion in human bronchial epithelial cells. Biochem. J. 2002, 367, 751–760. [Google Scholar] [CrossRef] [PubMed]
- Corrotte, M.; Chasserot-Golaz, S.; Huang, P.; Du, G.; Ktistakis, N.T.; Frohman, M.A.; Vitale, N.; Bader, M.F.; Grant, N.J. Dynamics and function of phospholipase D and phosphatidic acid during phagocytosis. Traffic 2006, 7, 365–377. [Google Scholar] [CrossRef] [PubMed]
- Antoine, J.C.; Prina, E.; Lang, T.; Courret, N. The biogenesis and properties of the parasitophorous vacuoles that harbour Leishmania in murine macrophages. Trends Microbiol. 1998, 6, 392–401. [Google Scholar] [CrossRef]
- Araki, N.; Hatae, T.; Furukawa, A.; Swanson, J.A. Phosphoinositide-3-kinase-independent contractile activities associated with Fcgamma-receptor-mediated phagocytosis and macropinocytosis in macrophages. J. Cell Sci. 2003, 116, 247–257. [Google Scholar] [CrossRef] [PubMed]
- Pridgeon, J.W.; Olzmann, J.A.; Chin, L.S.; Li, L. PINK1 protects against oxidative stress by phosphorylating mitochondrial chaperone TRAP1. PLoS Biol. 2007, 5, e172. [Google Scholar] [CrossRef] [PubMed]
- Mishra, S.; Fujita, T.; Lama, V.N.; Nam, D.; Liao, H.; Okada, M.; Minamoto, K.; Yoshikawa, Y.; Harada, H.; Pinsky, D.J. Carbon monoxide rescues ischemic lungs by interrupting MAPK-driven expression of early growth response 1 gene and its downstream target genes. Proc. Natl. Acad. Sci. USA 2006, 103, 5191–5196. [Google Scholar] [CrossRef] [PubMed]
- Reed, J.C.; Doctor, K.; Rojas, A.; Zapata, J.M.; Stehlik, C.; Fiorentino, L.; Damiano, J.; Roth, W.; Matsuzawa, S.; Newman, R.; et al. Comparative analysis of apoptosis and inflammation genes of mice and humans. Genome Res. 2003, 13, 1376–1388. [Google Scholar] [CrossRef] [PubMed]
- Diefenbach, A.; Schindler, H.; Donhauser, N.; Lorenz, E.; Laskay, T.; MacMicking, J.; Rollinghoff, M.; Gresser, I.; Bogdan, C. Type 1 interferon (IFNα/β) and type 2 nitric oxide synthase regulate the innate immune response to a protozoan parasite. Immunity 1998, 8, 77–87. [Google Scholar] [CrossRef]
- Zhou, J.; Fandrey, J.; Schumann, J.; Tiegs, G.; Brune, B. No and TNF-α released from activated macrophages stabilize HIF-1α in resting tubular LLC-PK1 cells. Am. J. Physiol. Cell Physiol. 2003, 284, C439–C446. [Google Scholar] [CrossRef] [PubMed]
- Yan, H.; Tsai, M.D. Nucleoside monophosphate kinases: Structure, mechanism, and substrate specificity. Adv. Enzymol. Relat. Areas Mol. Biol. 1999, 73, 103–134. [Google Scholar] [PubMed]
- Villa, H.; Perez-Pertejo, Y.; Garcia-Estrada, C.; Reguera, R.M.; Requena, J.M.; Tekwani, B.L.; Balana-Fouce, R.; Ordonez, D. Molecular and functional characterization of adenylate kinase 2 gene from Leishmania donovani. Eur. J. Biochem. 2003, 270, 4339–4347. [Google Scholar] [CrossRef] [PubMed]
- Goldman-Pinkovich, A.; Balno, C.; Strasser, R.; Zeituni-Molad, M.; Bendelak, K.; Rentsch, D.; Ephros, M.; Wiese, M.; Jardim, A.; Myler, P.J.; et al. An arginine deprivation response pathway is induced in Leishmania during macrophage invasion. PLoS Pathog. 2016, 12, e1005494. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Clarencio, J.; de Oliveira, C.I.; Bomfim, G.; Pompeu, M.M.; Teixeira, M.J.; Barbosa, T.C.; Souza-Neto, S.; Carvalho, E.M.; Brodskyn, C.; Barral, A.; et al. Characterization of the T-cell receptor Vβ repertoire in the human immune response against Leishmania parasites. Infect. Immun. 2006, 74, 4757–4765. [Google Scholar] [CrossRef] [PubMed]
- Keesen, T.S.; Antonelli, L.R.; Faria, D.R.; Guimaraes, L.H.; Bacellar, O.; Carvalho, E.M.; Dutra, W.O.; Gollob, K.J. CD4+ T cells defined by their Vβ T cell receptor expression are associated with immunoregulatory profiles and lesion size in human leishmaniasis. Clin. Exp. Immunol. 2011, 165, 338–351. [Google Scholar] [CrossRef] [PubMed]
- Lord, S.J.; Rajotte, R.V.; Korbutt, G.S.; Bleackley, R.C. Granzyme B: A natural born killer. Immunol. Rev. 2003, 193, 31–38. [Google Scholar] [CrossRef] [PubMed]
- Pucadyil, T.J.; Tewary, P.; Madhubala, R.; Chattopadhyay, A. Cholesterol is required for Leishmania donovani infection: Implications in leishmaniasis. Mol. Biochem. Parasitol. 2004, 133, 145–152. [Google Scholar] [CrossRef] [PubMed]
- Ben-Othman, R.; Flannery, A.R.; Miguel, D.C.; Ward, D.M.; Kaplan, J.; Andrews, N.W. Leishmania-mediated inhibition of iron export promotes parasite replication in macrophages. PLoS Pathog. 2014, 10, e1003901. [Google Scholar] [CrossRef] [PubMed]
- Chava, A.K.; Chatterjee, M.; Sharma, V.; Sundar, S.; Mandal, C. Variable degree of alternative complement pathway-mediated hemolysis in Indian visceral leishmaniasis induced by differential expression of 9-O-acetylated sialoglycans. J. Infect. Dis. 2004, 189, 1257–1264. [Google Scholar] [CrossRef] [PubMed]
- Candiano, G.; Bruschi, M.; Musante, L.; Santucci, L.; Ghiggeri, G.M.; Carnemolla, B.; Orecchia, P.; Zardi, L.; Righetti, P.G. Blue silver: A very sensitive colloidal coomassie G-250 staining for proteome analysis. Electrophoresis 2004, 25, 1327–1333. [Google Scholar] [CrossRef] [PubMed]
- Yan, J.X.; Wait, R.; Berkelman, T.; Harry, R.A.; Westbrook, J.A.; Wheeler, C.H.; Dunn, M.J. A modified silver staining protocol for visualization of proteins compatible with matrix-assisted laser desorption/ionization and electrospray ionization-mass spectrometry. Electrophoresis 2000, 21, 3666–3672. [Google Scholar] [CrossRef]
- Friedman, D.B.; Wang, S.E.; Whitwell, C.W.; Caprioli, R.M.; Arteaga, C.L. Multivariable difference gel electrophoresis and mass spectrometry: A case study on transforming growth factor-β and ERBB2 signaling. Mol. Cell. Proteom. 2007, 6, 150–169. [Google Scholar] [CrossRef] [PubMed]
- Martinez-Subiela, S.; Bernal, L.J.; Ceron, J.J. Serum concentrations of acute-phase proteins in dogs with leishmaniosis during short-term treatment. Am. J. Vet. Res. 2003, 64, 1021–1026. [Google Scholar] [CrossRef] [PubMed]
- Wasunna, K.M.; Raynes, J.G.; Were, J.B.; Muigai, R.; Sherwood, J.; Gachihi, G.; Carpenter, L.; McAdam, K.P. Acute phase protein concentrations predict parasite clearance rate during therapy for visceral leishmaniasis. Trans. R. Soc. Trop. Med. Hyg. 1995, 89, 678–681. [Google Scholar] [CrossRef]
- Cavalcanti, A.S.; Ribeiro-Alves, M.; Pereira Lde, O.; Mestre, G.L.; Ferreira, A.B.; Morgado, F.N.; Boite, M.C.; Cupolillo, E.; Moraes, M.O.; Porrozzi, R. Parasite load induces progressive spleen architecture breakage and impairs cytokine mRNA expression in Leishmania infantum-naturally infected dogs. PLoS ONE 2015, 10, e0123009. [Google Scholar] [CrossRef] [PubMed]



© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Veras, P.S.T.; Bezerra de Menezes, J.P. Using Proteomics to Understand How Leishmania Parasites Survive inside the Host and Establish Infection. Int. J. Mol. Sci. 2016, 17, 1270. https://doi.org/10.3390/ijms17081270
Veras PST, Bezerra de Menezes JP. Using Proteomics to Understand How Leishmania Parasites Survive inside the Host and Establish Infection. International Journal of Molecular Sciences. 2016; 17(8):1270. https://doi.org/10.3390/ijms17081270
Chicago/Turabian StyleVeras, Patrícia Sampaio Tavares, and Juliana Perrone Bezerra de Menezes. 2016. "Using Proteomics to Understand How Leishmania Parasites Survive inside the Host and Establish Infection" International Journal of Molecular Sciences 17, no. 8: 1270. https://doi.org/10.3390/ijms17081270
APA StyleVeras, P. S. T., & Bezerra de Menezes, J. P. (2016). Using Proteomics to Understand How Leishmania Parasites Survive inside the Host and Establish Infection. International Journal of Molecular Sciences, 17(8), 1270. https://doi.org/10.3390/ijms17081270