1. Introduction
As sessile organisms, plants frequently suffer from adverse environmental factors in their lifetimes. Of the various environmental stresses, drought is one of the factors that negatively affects plant growth and development, as well as crop yields and quality. With the ever-increasing world population, rapid development of the economy, and global climate change, the semi-arid and arid regions, worldwide, cover a substantial portion of the land area [
1]. Thus, it is of significance to elucidate the plant responses to drought stress and the underlying molecular mechanisms.
To survive and complete lifecycles in the stressful environment, plants have evolved a series of defense and adaptation mechanisms to reduce the damage caused by the water deficiency stress. In recent years, extensive studies using transcriptomic and proteomic approaches have been performed to investigate the plant responses to drought stress, and these works led to the identification of a large number of stress-responsive genes/proteins [
2,
3]. Further characterization of these stress-responsive genes/proteins promotes our understanding of the abiotic stress regulatory networks in plants. Now, it is commonly accepted that upon perception of the stress signal by some stress sensors, the stress signal may be transmitted via multiple signal cascades, including the mitogen-activated protein kinase (MAPK) pathway. Reactive oxygen species (ROS) and Ca
2+ are believed to function as two important signaling molecules in stress signaling and plant hormones, in particular, abscisic acid (ABA), also play crucial roles in stress signaling. Stress signaling in plants results in extensive biochemical and physiological alteration, as well as gene regulation.
Protein phosphorylation is one of the most important post-translational protein modifications in cells, and the reversible phosphorylation of proteins regulates many aspects of cell activities [
4]. In particular, research showed that protein phosphorylation modification plays an important role in the response to abiotic stress by regulating stomatal switching [
5], and defense and stress response in plants [
4]. In recent years, large-scale protein phosphorylation analyses have been performed to investigate the protein phosphorylation-medicated abiotic stress response in rice [
6],
Arabidopsis thaliana [
7], maize [
8],
Brachypodium distachyon [
9], and wheat [
10]. However, these phosphoproteomic analyses were performed in model plants and crops, and far less study has focused on non-model plants.
Ammopiptanthus mongolicus, an ecologically important shrub in mid-Asia desert regions, exhibits high levels of drought and low-temperature tolerances and is a rare evergreen broad-leafed shrub in the northwest desert of China.
A. mongolicus has been increasingly used as a model in abiotic tolerance research in trees [
11]. The transcriptome analyses of drought stress responses, and miRNA identification and profiling under drought stress, were carried out in
A. mongolicus [
11,
12,
13,
14]. These studies promote our understanding of the molecular mechanisms underlying the drought stress response in
A. mongolicus and provide important nucleotide and amino acid sequence data for study at the protein level in
A. mongolicus.
The root is the organ of the plant that takes up water and nutrients from the soil. At the same time, the root probably plays a key role in stress perception, signal transduction, and stress response in plants under drought stress. In the present study, we performed a quantitative proteomic analysis of the response of A. mongolicus roots under drought stress, using TiO2 enrichment and Liquid chromatography–tandem mass spectrometry (LC–MS/MS) analysis, coupled with Isobaric tags for relative and absolute quantitation (iTRAQ) labeling quantification. A large number of phosphorylation sites and phosphoproteins involved in the drought stress response were identified. These drought-corresponsive phosphoproteins, and the related signaling and metabolic pathways, probably play important roles in drought stress signaling and response in A. mongolicus roots.
2. Materials and Methods
2.1. Material and Stress Treatment
The seeds of A. mongolicus were collected in Zhongwei city, Ningxia autonomous district, China. The seeds were surface sterilized using 70% (v/v) ethanol for 1 min, followed by bleaching (10%) for 6 min, and then rinsed with distilled water thoroughly and germinated on moistened filter paper for 48 h at 25 °C. The seeds were planted in a 30 cm diameter pot containing a 3:1 (v/v) mixture of vermiculite and perlite. Seedlings were grown in a greenhouse under 120 μmol m−2 s−1 photosynthetic photon flux density, with a photoperiod of 16 h light and 8 h dark cycle, at approximately 25 °C and 35% relative humidity. The seedlings were watered every four days with half-length Hoagland solution. Eight weeks after germination, the seedlings were subjected to drought treatment.
For drought stress treatments, the seedlings were randomly divided into two groups. The drought treated group was irrigated with 20% PEG-6000 for 1 h, whereas the unstressed group was used as the control. All root samples of the two groups were collected, snap-frozen in nitrogen, and stored at −80 °C until further processing. Four biological replicates were used for iTRAQ labeling in the control or drought stress-treated group, and each biological replicate in the control or drought stress-treated group was pooled from 5 to 8 seedlings.
2.2. Protein Extraction
Approximately 0.5 g of fresh root from each biological replicate were ground into a fine power in liquid nitrogen in a mortar. The power was transferred into an acetone solution containing 10% trichloroacetic acid and 65 mM Dithiothreitol (DTT) in a 50 mL tube, and incubated at −20 °C for 1 h. After centrifugation at 17,000× g for 45 min, the resulting supernatant was discarded and the pellet was air-dried. The pellet was then suspended in STD buffer (4% SDS, 1 mM DTT, 150 mM Tris-HCl, pH 8.0) and mixed thoroughly by vortexing. After 3 min incubation in boiling water, the suspension was sonicated, incubated in boiling water for another 5 min and then clarified by centrifugation. The protein concentration was determined by the bicinchoninic acid protein assay kit (Beyotime, Beijing, China). The quality of the protein samples were further checked by Sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) electrophoresis.
2.3. Protein Digestion and iTRAQ Labeling
Protein digestion was performed according to the filter-aided sample preparation (FASP) procedure described by Wisniewki et al. [
15] and Hu et al. [
16]. The peptide content was estimated by spectra density using UV absorption at 280 nm. The resulting peptide mixture was labeled using the 8-plex iTRAQ reagent according to the manufacturer’s instructions (AB SCIEX) (113 to 116 tags for the four biological replicates of the control group, 117 to 121 tags for the four biological replicates of the drought stress-treated group) and vacuum dried.
2.4. Enrichment of Phosphorylated Peptiedes by the TiO2 Beads
The vacuum-dried peptide mixture was re-suspended in 1× DHB buffer, then added to the appropriate amount of TiO2 beads and agitated for 40 min. The sample was centrifuged for 1 min at 5000× g, resulting in the first set of beads. The supernatant from the first centrifugation was mixed with more TiO2 beads, which were treated before, resulting in the second set of beads. Both sets of beads were pooled and washed three times with wash buffer I (30% ACN/3%TFA, 50 μL) and then three times with washing buffer II (80% ACN/0.3% TFA, 50 μL). Finally, the phosphopeptides were eluted with elution buffer (40% ACN/15% NH4OH), lyophilized and re-suspended in 20 μL 0.1% formic acid. Five microliter peptide solution was used for mass spectral analysis.
2.5. Mass Spectrometry
Peptide separation was performed with an automated Easy-nLC1000 system coupled to a Q-Exactive mass spectrometer (Thermo Finnigan, San Jose, CA, USA). The enriched phosphopeptides were loaded onto a Thermos Scientific (Rockford, IL, USA) EASY columm (2 cm long, 100 μm inner diameter, RP-C18, 5 μm) equilibrated with 95% Buffer A (Buffer A, 0.1% formic acid), and then the peptides were loaded and separated on a C18 column (75 μm long, 250 mm inner diameter, RP-C18, 3 μm) at a flow rate of 250 nL/min. The peptides were eluted with a gradient of 0–55% buffer B (0.1% formic acid in acetonitrile) from 0 to 220 min, 55–100% buffer B from 220 to 228 min, and 100% buffer B from 228 to 240 min.
For MS data acquisition, the peptides were analyzed in positive ion mode at a resolution of 70,000 (at 200 m/z). The data-dependent top 10 MS/MS were selected by dynamically choosing the most abundant precursor ions from the survey scan (300–1800 m/z) using high-energy collision dissociation (HCD). Determination of the target value is based on predictive automatic gain control (pAGC). The dynamic exclusion duration was 25.0 s. Survey scans were acquired at a resolution of 17,500 at m/z 200. Normalized collision energy was 29 eV and the under-fill ratio, which specifies the minimum percentage of the target value likely to be reached at maximum-fill time, was defined as 0.1%. The instrument was run with peptide recognition mode enabled.
2.6. Data Analysis
Raw MS/MS data were interpreted with Proteome Discoverer (version 1.4, Thermo Fisher Scientific, Waltham, MA, USA) using the search engine Mascot (Version 2.2, Matrix Science, London, UK) against the protein database of
A. mongolicus reported previously [
14] and the decoy database. The Mascot parameters were set as follows: enzyme, trypsin; mass values, monoisotopic; peptide mass tolerance, ±20 ppm; MS/MS tolerance, 0.1 Da; max missed cleavages, 2; fixed modifications, carbamidomethyl (C), iTRAQ8plex (N-term), iTRAQ8plex (K); variable modifications, oxidation (M), phospho (ST), and phospho (Y). The decoy database pattern was set as the reverse of the target database. All reported data were based on 99% confidence for peptide identifications as determined by a false discovery rate (FDR) of ≤0.01. Protein identification was supported by at least one unique peptide identification. Phospho RS scores above 50 and PhosphoRS site probabilities above 75% indicate that a site is truly phosphorylated [
17].
2.7. Bioinformatics
The biological functions of the identified proteins were classified according to their gene ontology annotations and their annotation in KEGG (Kyoto Encyclopedia of Genes and Genomes) database. Protein-protein interaction networks were analyzed using the program STRING (
http://string-db.org/), a database of known and predicted protein-protein interactions. The confidence score was set at the medium level (≥0.400).
For conservation analysis of the phosphorylation sites in the identified differentially-phosphorylated phosphoproteins (DPPs), sequences of the DPPs were used as queries to BLAST search the
Arabidopsis protein database TAIR10 (
www.arabidopsis.org) to determine the
Arabidopsis orthologs, then the phosphorylation information of the corresponding S, T, and Y sites in PhosPhAt database (v4.0) [
18] were checked manually to determine the extent of the conservation of phosphorylation sites between ortholog pairs from
A. thaliana and
A. mongolicus. The thresholds for BLAST were set at an
E-value < 1 × 10
−10, and an identity ≥30%.
Motif-x [
19] was adapted for the identification of the phosphorylation motifs present in the phophoproteins. We extracted all items of P(S/T)P (6, 6), a phosphopeptide with a length of 13 with six upstream residues and six downstream residues surrounding the p-site. If the p-site was located in the N-terminus or C-terminus of the protein sequence, the phosphopeptide was complemented to P(S/T)P (6, 6) with the necessary number of “*”s instead of any amino acid. Motif-x parameter settings of “pre-aligned”, central S, T, or Y, width = 13, occurrence = 20, and significance = 0.000001 were adopted.
For the analysis of the structure preferences for the p-sites, NetSurfP ver. 1.1 (
www.cbs.dtu.dk) [
20] with the default parameters was adopted for the prediction of the secondary structures.
2.8. Western Blot and qRT-PCR
Western blot was conducted after 12% SDS-PAGE by probing with primary antibodies at the recommended dilutions: anti-HSP70 (1:1000, rabbit polyclonal; Beijing Protein Innovation, Beijing, China), anti-MPK6, and anti-PIP1 (1:2000, rabbit polyclonal; Beijing Protein Innovation, Beijing, China). Fifty to eighty micrograms of proteins were used depending on the sensitivity of the specific antibody. The protein concentration was measured using Bradford methods (Bioteke, Beijing, China). Goat anti-rabbit IgG (H+L) 800 CW (1:5000, LI-COR) was used as the secondary antibody. Visualization and quantification were carried out with the LI-COR Odyssey scanner and software (LI-COR Biosciences).
Quantitative real-time RT-PCR was performed according to a previously described protocol [
14]. All primers used in this study are listed in
Table S1 AmeIF1 (GenBank accession no. JN885965) was used as the internal reference gene.
2.9. Statistical Analysis
The phosphoprotein assays were performed in four biological replicates. Statistical analyses of phosphopeptides were conducted using the Student’s t-test with Microsoft Excel (version 2010, Microsoft Corp., Redmond, WA, USA) and a p-value ≤0.05 was considered statistically significant.
4. Discussion
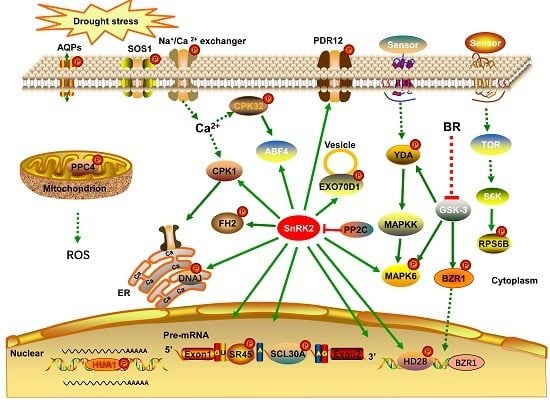
In the present study, we conducted an iTRAQ-based quantitative phosphoproteomic analysis of the response to short-term drought stress in A. mongolicus roots. A large number of phosphorylation sites were identified and a batch of phosphoproteins possibly involved in the drought stress response were found. These drought-responsive phosphoproteins and the related signaling and metabolic pathways may play important roles in drought stress signaling and response in A. mongolicus roots.
4.1. Phosphoproteins Involved in Signal Transduction and Transcriptional Regulation
Protein phosphorylation play a crucial role in signal transduction in plant cell. In the present study, eight kinases were found to be differentially phosphorylated under drought stress and these kinases probably participated in the drought stress signal transduction process in A. mongolicus roots.
ABA, a plant hormone, is a key signaling molecule regulating plant responses to abiotic stress. As expected, the motif analysis revealed that many phosphorylation sites in DPPs were binding sites of SnRK2, the key kinases of ABA signaling pathways, highlighting the role of the ABA signaling in drought stress response in A. mongolicus roots.
The MAPK cascade is one of the most important signaling pathways working in transmitting cellular signals. The MAPK cascade consists of three tier components: mitogen-activated protein kinase kinase kinases (MAPKKKs), mitogen-activated protein kinase kinases (MAPKKs), and mitogen-activated protein kinases (MAPKs), and the pathway carries out the phosphorylation reaction from the upstream receptor to the downstream target [
24]. Two kinases related to MAPK cascades, i.e., YDA and MAPK6, were differentially phosphorylated under drought stress in the present study, indicating that the MAPK pathway is involved in drought stress signaling in
A. mongolicus roots. YDA has been demonstrated to act upstream of the MKK4/MKK5-MPK3/MPK6 module to regulate stomatal development [
25] and, in a recent study, YDA was shown to be regulated at the transcriptional level by AN3 to improve root systems under drought stress [
26].
Ca
2+ participates in signal transduction in plants under environmental stress and calcium-dependent protein kinase (CDPKs) plays a crucial role in the Ca
2+-mediated signaling pathway. In the present study, The phosphorylation levels of CPK32 (
comp30972_c0_seq1) and a CDPK (
comp9850_c0_seq1) were down-regulated under drought stress, indicating the Ca
2+-mediated pathway participates in drought stress signaling in
A. mongolicus roots. CPK32 regulates the ABA-responsive gene expression via phosphorylation of the ABA-responsive element (ABRE)-binding transcription factors 4 (ABF4) in
Arabidopsis [
27].
Brassinosteroids (BRs), a group of plant steroid hormones, play important roles in plant growth, development, and stress responses. BR signaling through transmembrane receptor kinases and intracellular cascades leads to dephosphorylation and accumulation of the BZR1 [
28]. We found a BZR1 homolog (
comp7746_c0_seq1), the key transcription factor in the brassinosteroid (BR) signaling pathway were differentially phosphorylated under drought stress in
A. mongolicus roots. The down-regulated phosphopeptide, ISNs(ph)APVt(ph)PPLs(ph)SPTWR, harboring a typical motif of GSK-3 kinase, the key kinase mediating the BR response by phosphorylate BZR1. This observation suggested that the activity or abundance of GSK-3 may be decreased under drought stress.
In addition to BZR1, some other transcription factors (TFs) were found to be differentially phosphorylated under drought stress, for example, floral homeotic protein (HUA1) was involved in flower development under stressful environments [
29]. These TFs may contribute greatly to the gene regulation network in responding to the short-term drought stress.
4.2. Phosphoproteins Involved in Transmembrane Transport, Cytoskeleton, and Vesicle Transport
Maintenance of the cell’s osmotic potential under water-deficient conditions is a major challenge for plant growth and development. In the present study, several aquaporins and ion transporters were found to be differentially phosphorylated under drought stress, suggesting protein phosphorylation of these proteins may play roles in regulating osmotic balance upon short-term drought stress in A. mongolicus roots.
Aquaporins (AQPs), a family of highly-conserved transmembrane proteins, are involved in the quick transport of water and other small non-polar molecules, and regulation of the osmotic balance. We found a serine phosphorylation site in an up-regulated phosphopeptide: EQDVS(ph)LGANK in a PIP1-1 homolog (
comp29561_c0_seq1) under drought stress. PIPs were the most abundant aquaporins in the plasma membrane, and most PIPs are expressed dominantly in the root and down-regulated by water stress in
Arabidopsis [
30]. In addition to the transcriptional regulation, PIP1 and PIP2 subgroups were also regulated by phosphorylation and proper phosphorylation can modulate the channel activity of PIPs [
31]. We speculated that the differential phosphorylation upon drought stress might be a quick way to regulate activities of aquaporins in
A. mongolicus roots.
Several molecular pumps were also found to be differentially phosphorylated under drought stress in
A. mongolicus roots and some of them may contribute to the osmotic adjustment under drought stress. Among these proteins, SOS1 is one of the well-documented phosphoproteins, which is involved in ion transmembrane transport regulated by the SOS signaling pathway [
32]. Some of these pumps transport ions, such as sodium (SOS1), calcium (sodium/calcium exchanger family protein), and potassium (high-affinity K
+ transporter 5); the others transport amino acids and other small molecules (transmembrane amino acid transporter family protein, pleiotropic drug resistance 12, ABC-2 type transporter family protein, and tonoplast monosaccharide transporter 2). The alteration in phosphorylation status of these transporters upon drought treatment indicate their possible roles in regulating ions and small molecule concentrations to reduce water loss from cytoplasm. Such a regulation may not only contribute to osmotic adjustment and nutrient absorption, but also be associated with regulation of Ca
2+-mediated signaling.
Under the high osmotic environment, water efflux was enhanced and the cell volume became smaller in plant roots [
33]. To cope with such stress, the cytoskeleton, cell wall, and plasma membrane would change correspondingly. We found five cytoskeleton-related proteins differentially phosphorylated under drought stress. Of these phosphoproteins, four were related to microfilaments, including villin 2, villin 4, and formin homology 2, and the other phosphoprotein was related to microtubules. Phosphorylation modification of villins and formins were proposed to play roles in modulating their activities in human and animal cells [
34,
35]. In the present study, the cytoskeleton-related DPPs probably participate in modulating the cytoskeleton organization in drought-stressed root cells. The cell wall is critical for cell shape maintenance and two proteins involved in cell wall formation were regulated at the phosphorylation level. Of these proteins, the cellulose synthase family protein participate in the secondary cell wall formation, and glucan synthase-like 5 plays a role in callose deposition in the cell wall [
36].
In addition, five proteins involved in exocytosis and endocytosis were found to be differentially phosphorylated under drought stress. Previous study showed that short-term extracellular osmotic treatments result in a shift in the balance between endocytosis and exocytosis in root meristem cells by influencing the tension on the plasma membrane [
37], and we speculated that these five DPPs might contribute to the drought stress-induced alteration in tension on the plasma membrane in
A. mongolicus roots.
4.3. Phosphoproteins Involved in RNA Splicing and Transport
To create translatable mRNAs, pre-mRNA molecules need to be properly processed, spliced, and transported to the cytoplasm. Recent studies showed that post-transcriptional regulation, especially alternative splicing (AS) and mRNA export, is essential for plants to adapt to environmental stress. In the present study, a large number of phosphoproteins involved in the post-transcriptional regulation of RNA were identified, highlighting their important roles in drought stress response in A. mongolicus roots.
The SR (serine arginine-rich) proteins are a class of proteins participating in spliceosome assembly in plants. SR proteins consist of one or two RNA recognition motifs (RRMs) that contact the RNA targets and a domain enriched in serine and arginine residues involved in protein-protein interaction. We found three SR proteins whose phosphorylation levels were up-regulated under drought stress, and these SR proteins include SCL30A, SC35, and SR45a. In addition, there are four additional DPPs involved in RNA splicing and the phosphorylation levels of most of these phosphoproteins were increased under drought stress. Phosphorylation is proposed to modulate protein–protein interactions within the spliceosome, thereby contributing to dynamic structural reorganization of the spliceosome during splicing [
38]. Given that accumulating evidence supports the idea that AS is a new means of regulating the environmental fitness of plants [
39], we speculate that phosphorylation of RNA splicing-related proteins possibly serve as an immediate means to regulate the environmental fitness of plants by modulating the AS process in
A. mongolicus roots under short-term drought stress.
4.4. Phosphoproteins Involved in Protein Synthesis, Folding, and Degradation
Protein synthesis and folding may be disturbed and, hence, protein degradation may be activated in plants under stress condition, and previous proteomic studies have identified a large number of drought-corresponsive proteins associated with protein synthesis, folding, and degradation [
40]. In the present study, two DPPs involved in ribosomes and translation, four DPPs related to protein folding, and four DPPs associated with protein degradation were identified, suggesting the important role of protein phosphorylation in regulating protein turnover at the post-transcriptional level.
Ribosomal protein S6 (RPS6), a component of the 40 S ribosomal subunit, has been shown to be a key component of the TOR-S6k- RPS6 signaling pathway [
41]. Environmental cues, such as osmotic stress, affect phosphorylation status of RPS6 via TOR pathway and the phosphorylation level of RPS6 is closely associated with cell proliferation activity and cell size [
42]. The addition of fresh medium to
Arabidopsis suspension cell culture promotes phosphorylation of RPS6 by activating the cognate kinase of RPS6, AtS6k. In the present study, six serine phosphorylation sites were identified in an RPS6 homolog (
comp8048_c0_seq1) and the phosphorylation levels of these phosphorylation sites were down-regulated under drought stress. We speculate that the drought stress signal transmitted by the TOR pathway resulted in down-regulation of RPS6 phosphorylation observed in the present study and the alteration in RPS6 phosphorylation status may influence the root under drought stress.
Heat shock proteins (HSPs) are a family of proteins that are generated by cells upon exposure to stressful conditions and many members of HSPs perform chaperone functions. In the present study, two HSP20-like proteins (
comp41212_c0_seq1 and
comp17846_c0_seq1) were differentially phosphorylated under drought stress. The stress-induced change in phosphorylation status in small molecular HSPs was also observed in maize [
16].
In addition, four proteins involved in protein ubiquitination, and misfolded proteins exporting from the endoplasmic reticulum were found to be differentially phosphorylated under drought stress, indicating protein degradation via the ubiquitination pathway may be regulated by protein phosphorylation upon short-term drought stress in A. mongolicus roots. In brief, our study provided data to advance our understanding of the important role of protein phosphorylation in regulating protein turnover at the post-transcriptional level.
4.5. Phosphoproteins Involved in Epigenetic Regulation
Epigenetic regulations such as histone modification have been proposed to play key roles in gene regulation in plants under stress [
43]. Not surprisingly, three proteins involved in epigenetic regulations were differentially phosphorylated under drought stress in
A. mongolicus roots. Of these proteins, histone deacetylation mediate transcriptional repression and histone deacetylase 2B (HD2B) may play an important roles in transcriptional regulation in plant [
44].
4.6. Phosphoproteins Involved in Pyruvate Metabolism
Two enzymes involved in pyruvate metabolism were differentially phosphorylated under drought stress in
A. mongolicus roots. Phosphoenolpyruvate carboxylase 4 (PPC4, comp36964_c0_seq1) may provide more oxaloacetate used for the synthesis of amino acids, like aspartic acid and glutamic acid, which were observed to accumulate in plant cells under drought stress [
45]. PPC4 and malate dehydrogenase (
comp20184_c0_seq1) can work together to synthesize malate from glycolytically-derived phosphoenolpyruvate.
4.7. Gene Expression and Protein Abundance of Selected DPPs under Drought Stress
The alteration in phosphorylation level of an individual protein might have resulted from either the up-regulation of the activity or abundance of the corresponding kinase, or the increase of the protein abundance. The Western blots showed that although the phosphorylation level of MPK6 and PIP1 were up-regulated under drought stress, there were no significant alterations in protein abundance of the two proteins (
Figure S3). Thus, the drought stress-induced changes in phosphorylation status of MPK6 and PIP1 were probably caused by the alterations of the activity or abundance of the corresponding kinase.
Not surprisingly, the qRT-PCR analysis showed that drought stressed-induced changes in gene expression levels were not correlated with that of the phosphorylation status. However, it is noteworthy that the MPK6 and PIP1 encoding genes (
comp15811_c0_seq1 and
comp29561_c0_seq1) were observed to be up-regulated in transcription level (
Figure S4), which was different with that of protein abundance level. Many previous studies have reported the inconsistency between gene expression level and protein abundance level [
46], and our results indicated that DPPs, like MPK6 and PIP1, were regulated at both the transcription level and post-translational modification level during the first 1 h of drought stress in
A. mongolicus roots. Taken together, the difference between transcription level, protein accumulation level, and posttranslational level highlighted the complexity of the regulation network involved in plant response to environmental stresses.
5. Conclusions
Although extensive studies have been performed in investigating the molecular mechanism underlying the response to drought stress in plants, little is known about how reversible protein phosphorylation functions in the drought stress response. In the present study, we used an iTRAQ-based quantitative phosphoproteomic approach to analyze the short-term drought stress response in A. mongolicus, an ecologically important shrub in mid-Aisa desert regions. Of the large amount of phosphopeptides and phosphoproteins identified, 103 DEPPs and 90 DPPs were determined. The motif analysis of the phosphorylation sites in the DEPPs, together with functional and PPI analysis of the DPPs, revealed that signaling pathways, such as MAPK and CPK cascades, and ABA and BR hormone signaling pathways, were involved in stress signal transduction in A. mongolicus roots. Our results demonstrated that, as a rapid and reversible means to modulate protein activity, reversible protein phosphorylation mediated adjustment of diverse biological processes were induced by drought treatment, and these biological processes included osmotic adjustment, RNA splicing and export, protein synthesis, folding and degradation, as well as stress response and defense. Thus, this study provides important data for understanding the signal transduction and gene regulation mediated by reversible protein phosphorylation in plants in stressful conditions.