Recombinant Lipase from Gibberella zeae Exhibits Broad Substrate Specificity: A Comparative Study on Emulsified and Monomolecular Substrate
Abstract
:1. Introduction
2. Results and Discussion
2.1. Expression and Purification of Recombinant Gibberella zeae Lipase (rGZEL)
2.2. Biochemical Characterization of rGZEL under Classical Emulsion System
2.2.1. Effect of Temperature and pH on Various Lipolytic Activities of rGZEL
Lipase Activity
Phospholipase Activity
Glycolipid Hydrolysis Activity
2.2.2. Kinetic Studies of rGZEL to Different Substrates Using the Emulsion Method
2.2.3. Thermostability of rGZEL
2.2.4. Chain-Length Specificity of rGZEL
2.3. Kinetic Measurements by Monolayer Film Technique
2.3.1. Variations with Surface Pressure in Catalytic Activities Using Various Phospholipids as Substrate
2.3.2. Variations with Surface Pressure in Catalytic Activities Using Galactolipid as Substrate
2.3.3. Regioselectivity of rGZEL
2.3.4. Stereoselectivity of rGZEL
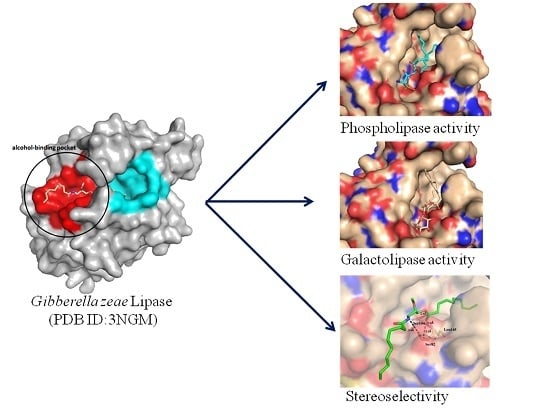
2.4. Docking of Phospholipid, Galactolipid and DDG Enantiomers into GZEL
3. Materials and Methods
3.1. Chemicals
3.2. Lipids
3.3. Vector Construction and Transformation of Escherichia coli Shuffle T7 Strain
3.4. Expression and Purification of Recombinant Enzyme
3.5. Biochemical Characterization of Recombinant Enzyme
3.5.1. Determinations of Lipolytic Activities Using Emulsified System
3.5.2. Determination of Temperature and pH Profiles
3.5.3. Determination of Kinetic Constants to Different Substrates
3.5.4. Determination on Chain-Length Specificity to Different Triglycerides (TAGs)
3.6. Monomolecular Film Experiments
3.6.1. Surface Pressure-Area Isotherms
3.6.2. Characterization on Kinetic Hydrolytic Activities of rGZEL to Different Substrates
3.7. Construction the Structures of GZEL with an Open Conformation and in Silico Docking
4. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
Abbreviations
| GZEL | Gibberella zeae Lipase |
| TC4 | Tributyrin |
| TC8 | Tricaprylin |
| TC18 | Triolein |
| PC | Phosphatidylcholine from Soybean |
| β-CD | β-Cyclodextrin |
| DOPC | 1,2-Dioleoyl-sn-Glycero-3-Phosphocholine |
| PI | l-α-Phosphatidylinositol |
| PS | 1,2-Diacyl-sn-Glycero-3-Phospho-l-Serine |
| PE | l-α-Phosphatidylethanolamine |
| CL | Cardiolipin |
| PA | 3-sn-Phosphatidic Acid Sodium Salt |
| SM | Sphingomyelin |
| PG | 1,2-Dioleoyl-sn-Glycero-3-Phospho-rac-(1-glycerol) Sodium Salt |
| MGDG | 1,2-Distearoyimonoglactosylglyceride |
| DDG | Didecanoyl-Deoxyamino-O-Methyl Glycerol |
| S1 | 1,2-Didecanoyl-2-Deoxyamino-3-O-Methyl Glycerol (1,2DDG), with the S Configuration |
| R1 | 2,3-Didecanoyl-2-Deoxyamino-1-O-Methyl Glycerol (2,3DDG), with the R Configuration |
| S2 | 1,3-Didecanoyl-3-Deoxyamino-2-O-Methyl Glycerol (1,3DDG), with the S Configuration |
| R2 | 1,3-Didecanoyl-1-Deoxyamino-2-O-Methyl Glycerol (1,3DDG), with the R Configuration |
| S3 | 2,3-Didecanoyl-3-Deoxyamino-1-O-Methyl Glycerol (2,3DDG), with the S Configuration |
| R3 | 1,2-Didecanoyl-1-Deoxyamino-3-O-Methyl Glycerol (1,2DDG), with the R Configuration |
| IPTG | Isopropyl β-d-1-Thiogalactopyranoside |
References
- Jaeger, K.E.; Dijkstra, B.W.; Reetz, M.T. Bacterial biocatalysts: Molecular biology, three-dimensional structures, and biotechnological applications of lipases. Annu. Rev. Microbiol. 1999, 53, 315–351. [Google Scholar] [CrossRef] [PubMed]
- Schmid, R.D.; Verger, R. Lipases: Interfacial enzymes with attractive applications. Angew. Chem. Int. Ed. 1998, 37, 1608–1633. [Google Scholar] [CrossRef]
- Bassegoda, A.; Pastor, F.I.J.; Diaz, P. Rhodococcus sp. strain CR-53 lipR, the first member of a new bacterial lipase family (family X) displaying an unusual Y-type oxyanion hole, similar to the Candida antarctica lipase clan. Appl. Environ. Microbiol. 2012, 78, 1724–1732. [Google Scholar] [CrossRef] [PubMed]
- Gupta, R.; Gupta, N.; Rathi, P. Bacterial lipases: An overview of production, purification and biochemical properties. Appl. Microbiol. Biotechnol. 2004, 64, 763–781. [Google Scholar] [CrossRef] [PubMed]
- Jaeger, K.E.; Eggert, T. Lipases for biotechnology. Curr. Opin. Biotechnol. 2002, 13, 390–397. [Google Scholar] [CrossRef]
- De Maria, L.; Vind, J.; Oxenboll, K.M.; Svendsen, A.; Patkar, S. Phospholipases and their industrial applications. Appl. Microbiol. Biotechnol. 2007, 74, 290–300. [Google Scholar] [CrossRef] [PubMed]
- Ramrakhiani, L.; Chand, S. Recent progress on phospholipases: Different sources, assay methods, industrial potential and pathogenicity. Appl. Biochem. Biotechnol. 2011, 164, 991–1022. [Google Scholar] [CrossRef] [PubMed]
- Voigt, C.A.; Schäfer, W.; Salomon, S. A secreted lipase of Fusarium graminearum is a virulence factor required for infection of cereals. Plant J. 2005, 42, 364–375. [Google Scholar] [CrossRef] [PubMed]
- Lou, Z.Y.; Li, M.; Sun, Y.; Liu, Y.; Liu, Z.; Wu, W.P.; Rao, Z.H. Crystal structure of a secreted lipase from Gibberella zeae reveals a novel “double-lock” mechanism. Protein Cell 2010, 1, 760–770. [Google Scholar] [CrossRef] [PubMed]
- Gutiérrez-Ayesta, C.; Carelli, A.A.; Ferreira, M.L. Relation between lipase structures and their catalytic ability to hydrolyse triglycerides and phospholipids. Enzyme Microb. Technol. 2007, 41, 35–43. [Google Scholar] [CrossRef]
- Xin, R.P.; Khan, F.I.; Zhao, Z.X.; Zhang, Z.D.; Yang, B.; Wang, Y.H. A comparative study on kinetics and substrate specificities of Phospholipase A1 with Thermomyces lanuginosus lipase. J. Colloid Interface Sci. 2017, 488, 149–154. [Google Scholar] [CrossRef] [PubMed]
- Yuan, D.J.; Zhao, Z.X.; Wang, X.M.; Guo, S.H.; Yang, B.; Wang, Y.H. Sequence-based proline incorporation improves the thermostability of Candida albicans lipase Lip5. Eur. J. Lipid Sci. Technol. 2016, 118, 821–826. [Google Scholar] [CrossRef]
- Aloulou, A.; Rodriguez, J.A.; Fernandez, S.; van Oosterhout, D.; Puccinelli, D.; Carrière, F. Exploring the specific features of interfacial enzymology based on lipase studies. Biochim. Biophys. Acta 2006, 1761, 995–1013. [Google Scholar] [CrossRef] [PubMed]
- Singh, H.; Ye, A.; Horne, D. Structuring food emulsions in the gastrointestinal tract to modify lipid digestion. Prog. Lipid Res. 2009, 48, 92–100. [Google Scholar] [CrossRef] [PubMed]
- Golding, M.; Wooster, T.J. The influence of emulsion structure and stability on lipid digestion. Curr. Opin. Colloid Interface Sci. 2010, 15, 90–101. [Google Scholar] [CrossRef]
- Calvez, P.; Bussiéres, S.; Demers, É.; Salesse, C. Parameters modulating the maximum insertion pressure of proteins and peptides in lipid monolayers. Biochimie 2009, 91, 718–733. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.H.; Zhang, H.; Liu, Y.H.; Khan, F.I.; Yang, B.; Wang, Y.H. Profiling substrate specificity of Lecitase Ultra to different kinds of phospholipids using monolayer technology. Eur. J. Lipid Sci. Technol. 2017, 119. [Google Scholar] [CrossRef]
- Horchani, H.; Ben Salem, N.; Chaari, A.; Sayari, A.; Gargouri, Y.; Verger, R. Staphylococcal lipases stereoselectively hydrolyse the sn-2 position of monomolecular films of diglyceride analogs. Application to sn-2 hydrolysis of triolein. J. Colloid Interface Sci. 2010, 347, 301–308. [Google Scholar] [CrossRef] [PubMed]
- Jallouli, R.; Othman, H.; Amara, S.; Parsiegla, G.; Carrière, F.; Srairi-Abid, N.; Gargouri, Y.; Bezzine, S. The galactolipase activity of Fusarium solani (phospho)lipase. Biochim. Biophys. Acta 2015, 1851, 282–289. [Google Scholar] [CrossRef] [PubMed]
- Siebertz, H.P.; Heinz, E.; Linscheid, M.; Joyard, J.; Douce, R. Characterization of lipids from chloroplast envelopes. Eur. J. Biochem. 1979, 101, 429–438. [Google Scholar]
- Amara, S.; Lafont, D.; Parsiegla, G.; Point, V.; Chabannes, A.; Rousset, A.; Carrière, F. The galactolipase activity of some microbial lipases and pancreatic enzymes. Eur. J. Lipid Sci. Technol. 2013, 115, 442–451. [Google Scholar] [CrossRef]
- Hoyo, J.; Guaus, E.; Torrentburgués, J. Monogalactosyldiacylglycerol and digalactosyldiacylglycerol role, physical states, applications and biomimetic monolayer films. Eur. Phys. J. 2016, 39, 39. [Google Scholar] [CrossRef] [PubMed]
- Deveer, A.M.; Dijkman, R.; Leuvelingtjeenk, M.; van den Berg, L.; Ransac, S.; Batenburg, M.; Egmond, M.; Verheij, H.M.; de Haas, G.H. A monolayer and bulk study on the kinetic behavior of Pseudomonas glumae lipase using synthetic pseudoglycerides. Biochemistry 1991, 30, 10034–10042. [Google Scholar] [CrossRef] [PubMed]
- Rogalska, E.; Nury, S.; Douchet, I.; Verger, R. Lipase stereoselectivity and regioselectivity toward three isomers of dicaprin: A kinetic study by the monomolecular film technique. Chirality 1995, 7, 505–515. [Google Scholar] [CrossRef]
- Douchet, I.; de Haas, G.; Verger, R. Lipase regio- and stereoselectivities toward three enantiomeric pairs of didecanoyl-deoxyamino-O-methyl glycerol: A kinetic study by the monomolecular film technique. Chirality 2003, 15, 220–226. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.G.; Wang, Y.H.; Yang, B.; Mainda, G.; Guo, Y. Degumming of vegetable oil by a new microbial lipase. Food Technol. Biotechnol. 2006, 44, 101–104. [Google Scholar]
- Verger, R.; de Haas, G.H. Enzyme reactions in a membrane model 1: A new technique to study enzyme reactions in monolayers. Chem. Phys. Lipids 1973, 10, 127–136. [Google Scholar] [CrossRef]






| Activity | pH | Temperature (°C) | Specific Activity (U/g) | Ratio |
|---|---|---|---|---|
| Lipase activity | 7.0 | 30.0–40.0 | 153,789.37 ± 776.75 | 1.00 |
| Phospholipase activity | 6.0–7.0 | 45.0 | 3698.95 ± 137.66 | 0.02 1 |
| Glycolipid hydrolysis activity | 6.0 | 50.0 | 9252.69 ± 633.15 | 0.06 2 |
| Substrate | Km (M) × 10−2 | Vmax (μM min−1 mg−1) | kcat (s−1) | kcat/Km (s−1 M−1) |
|---|---|---|---|---|
| Olive oil | 9.02 ± 0.07 | 171,67.38 ± 282.84 | 173.33 ± 1.14 | 1923.17 |
| Soybean-PC | 5.18 ± 0.03 | 1101.32 ± 14.12 | 11.12 ± 0.15 | 214.82 |
| Sucrose ester | 18.75 ± 3.25 | 5434.78 ± 21.21 | 54.88 ± 0.43 | 292.66 |
| Surface Pressure (mN/m) | Regioselectivity Index (VI) | Stereoselectivity Index (SI) | Ratio | ||
|---|---|---|---|---|---|
| [(A1,3 + A1,3) − (A1,2 + A2,3)]/[(A1,3 + A1,3) + (A1,2 + A2,3)] | (A2,3 − A1,2)/(A2,3 + A1,2) | (A1,3 − A1,3)/(A1,3 + A1,3) | (A1,2 − A2,3)/(A1,2 + A2,3) | [∣(A1,3 − A1,3)/(A1,3 + A1,3)∣]/[∣(A2,3 − A1,2)/(A2,3 + A1,2)∣] | |
| Primary ester with vicinal acyl chains | Primary ester with distal acyl chains | Secondary ester with vicinal acyl chains | |||
| 25 | +0.48 | −0.74 | +0.56 | 0 | 0.76 |
| 30 | +0.22 | +0.63 | +0.04 | 0 | 0.06 |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, F.; Zhang, H.; Zhao, Z.; Wei, R.; Yang, B.; Wang, Y. Recombinant Lipase from Gibberella zeae Exhibits Broad Substrate Specificity: A Comparative Study on Emulsified and Monomolecular Substrate. Int. J. Mol. Sci. 2017, 18, 1535. https://doi.org/10.3390/ijms18071535
Wang F, Zhang H, Zhao Z, Wei R, Yang B, Wang Y. Recombinant Lipase from Gibberella zeae Exhibits Broad Substrate Specificity: A Comparative Study on Emulsified and Monomolecular Substrate. International Journal of Molecular Sciences. 2017; 18(7):1535. https://doi.org/10.3390/ijms18071535
Chicago/Turabian StyleWang, Fanghua, Hui Zhang, Zexin Zhao, Ruixia Wei, Bo Yang, and Yonghua Wang. 2017. "Recombinant Lipase from Gibberella zeae Exhibits Broad Substrate Specificity: A Comparative Study on Emulsified and Monomolecular Substrate" International Journal of Molecular Sciences 18, no. 7: 1535. https://doi.org/10.3390/ijms18071535
APA StyleWang, F., Zhang, H., Zhao, Z., Wei, R., Yang, B., & Wang, Y. (2017). Recombinant Lipase from Gibberella zeae Exhibits Broad Substrate Specificity: A Comparative Study on Emulsified and Monomolecular Substrate. International Journal of Molecular Sciences, 18(7), 1535. https://doi.org/10.3390/ijms18071535