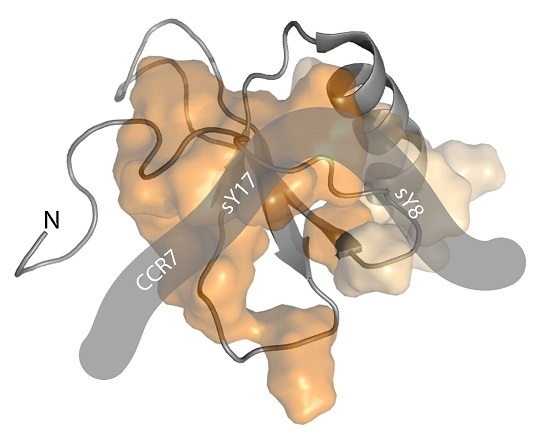
CCR7 Sulfotyrosine Enhances CCL21 Binding
Abstract
:1. Introduction
2. Results
2.1. A CCR7 N-Terminal Peptide Binds CCL21
2.2. CCR7 N-Terminal Peptides Maintain Binding Site Specificity upon Tyrosine Modification
2.3. Sulfotyrosine or Phosphotyrosine Modification Increase the Affinity of CCR7 N-Terminal Peptides for CCL21
3. Discussion
4. Materials and Methods
4.1 Recombinant Protein Purification and Peptide Synthesis
4.2 Protein NMR
Acknowledgments
Author Contributions
Conflicts of Interest
Abbreviations
| CCL21 | CC chemokine ligand 21 |
| CCL19 | CC chemokine ligand 19 |
| CCR7 | CC chemokine receptor 7 |
| NMR | Nuclear magnetic resonance |
| sY | Sulfotyrosine |
| pY | Phosphotyrosine |
References
- Baggiolini, M. Chemokines in pathology and medicine. J. Intern. Med. 2001, 250, 91–104. [Google Scholar] [CrossRef] [PubMed]
- Proudfoot, A.E. Chemokine receptors: Multifaceted therapeutic targets. Nat. Rev. Immunol. 2002, 2, 106–115. [Google Scholar] [CrossRef] [PubMed]
- Ben-Baruch, A. Organ selectivity in metastasis: Regulation by chemokines and their receptors. Clin. Exp. Metastasis 2008, 25, 345–356. [Google Scholar] [CrossRef] [PubMed]
- Wiley, H.E.; Gonzalez, E.B.; Maki, W.; Wu, M.T.; Hwang, S.T. Expression of CC chemokine receptor-7 and regional lymph node metastasis of B16 murine melanoma. J. Natl. Cancer Inst. 2001, 93, 1638–1643. [Google Scholar] [CrossRef] [PubMed]
- Oliveira-Neto, H.H.; de Souza, P.P.; da Silva, M.R.; Mendonca, E.F.; Silva, T.A.; Batista, A.C. The expression of chemokines CCL19, CCL21 and their receptor CCR7 in oral squamous cell carcinoma and its relevance to cervical lymph node metastasis. Tumour Biol. 2013, 34, 65–70. [Google Scholar] [CrossRef] [PubMed]
- Legler, D.F.; Uetz-von Allmen, E.; Hauser, M.A. CCR7: Roles in cancer cell dissemination, migration and metastasis formation. Int. J. Biochem. Cell Biol. 2014, 54, 78–82. [Google Scholar] [CrossRef] [PubMed]
- Moore, K.L. The biology and enzymology of protein tyrosine O-sulfation. J. Biol. Chem. 2003, 278, 24243–24246. [Google Scholar] [CrossRef] [PubMed]
- Moore, K.L. Protein tyrosine sulfation: A critical posttranslation modification in plants and animals. Proc. Natl. Acad. Sci. USA 2009, 106, 14741–14742. [Google Scholar] [CrossRef] [PubMed]
- Seibert, C.; Sanfiz, A.; Sakmar, T.P.; Veldkamp, C.T. Preparation and analysis of N-terminal chemokine receptor sulfopeptides using tyrosylprotein sulfotransferase enzymes. Methods Enzymol. 2016, 570, 357–388. [Google Scholar] [PubMed]
- Seibert, C.; Sakmar, T.P. Toward a framework for sulfoproteomics: Synthesis and characterization of sulfotyrosine-containing peptides. Biopolymers 2008, 90, 459–477. [Google Scholar] [CrossRef] [PubMed]
- Stone, M.J.; Chuang, S.; Hou, X.; Shoham, M.; Zhu, J.Z. Tyrosine sulfation: An increasingly recognised post-translational modification of secreted proteins. New Biotechnol. 2009, 25, 299–317. [Google Scholar] [CrossRef]
- Stone, M.J.; Payne, R.J. Homogeneous sulfopeptides and sulfoproteins: Synthetic approaches and applications to characterize the effects of tyrosine sulfation on biochemical function. Acc. Chem. Res. 2015, 48, 2251–2261. [Google Scholar] [CrossRef] [PubMed]
- Ludeman, J.P.; Stone, M.J. The structural role of receptor tyrosine sulfation in chemokine recognition. Br. J. Pharmacol. 2014, 171, 1167–1179. [Google Scholar] [CrossRef] [PubMed]
- Farzan, M.; Babcock, G.J.; Vasilieva, N.; Wright, P.L.; Kiprilov, E.; Mirzabekov, T.; Choe, H. The role of post-translational modifications of the cxcr4 amino terminus in stromal-derived factor 1 α association and HIV-1 entry. J. Biol. Chem. 2002, 277, 29484–29489. [Google Scholar] [CrossRef] [PubMed]
- Veldkamp, C.T.; Seibert, C.; Peterson, F.C.; Sakmar, T.P.; Volkman, B.F. Recognition of a CXCR4 sulfotyrosine by the chemokine stromal cell-derived factor-1α (SDF-1α/CXCL12). J. Mol. Biol. 2006, 359, 1400–1409. [Google Scholar] [CrossRef] [PubMed]
- Seibert, C.; Veldkamp, C.T.; Peterson, F.C.; Chait, B.T.; Volkman, B.F.; Sakmar, T.P. Sequential tyrosine sulfation of CXCR4 by tyrosylprotein sulfotransferases. Biochemistry 2008, 47, 11251–11262. [Google Scholar] [CrossRef] [PubMed]
- Millard, C.J.; Ludeman, J.P.; Canals, M.; Bridgford, J.L.; Hinds, M.G.; Clayton, D.J.; Christopoulos, A.; Payne, R.J.; Stone, M.J. Structural basis of receptor sulfotyrosine recognition by a CC chemokine: The N-terminal region of CCR3 bound to CCL11/eotaxin-1. Structure 2014, 22, 1571–1581. [Google Scholar] [CrossRef] [PubMed]
- Tan, J.H.; Ludeman, J.P.; Wedderburn, J.; Canals, M.; Hall, P.; Butler, S.J.; Taleski, D.; Christopoulos, A.; Hickey, M.J.; Payne, R.J.; et al. Tyrosine sulfation of chemokine receptor CCR2 enhances interactions with both monomeric and dimeric forms of the chemokine monocyte chemoattractant protein-1 (MCP-1). J. Biol. Chem. 2013, 288, 10024–10034. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.Z.; Millard, C.J.; Ludeman, J.P.; Simpson, L.S.; Clayton, D.J.; Payne, R.J.; Widlanski, T.S.; Stone, M.J. Tyrosine sulfation influences the chemokine binding selectivity of peptides derived from chemokine receptor CCR3. Biochemistry 2011, 50, 1524–1534. [Google Scholar] [CrossRef] [PubMed]
- Amarandi, R.M.; Hjorto, G.M.; Rosenkilde, M.M.; Karlshoj, S. Probing biased signaling in chemokine receptors. Methods Enzymol. 2016, 570, 155–186. [Google Scholar] [PubMed]
- Zidar, D.A.; Violin, J.D.; Whalen, E.J.; Lefkowitz, R.J. Selective engagement of G protein coupled receptor kinases (GRKs) encodes distinct functions of biased ligands. Proc. Natl. Acad. Sci. USA 2009, 106, 9649–9654. [Google Scholar] [CrossRef] [PubMed]
- Hofsteenge, J.; Stone, S.R.; Donella-Deana, A.; Pinna, L.A. The effect of substituting phosphotyrosine for sulphotyrosine on the activity of hirudin. Eur. J. Biochem. 1990, 188, 55–59. [Google Scholar] [CrossRef] [PubMed]
- Farzan, M.; Mirzabekov, T.; Kolchinsky, P.; Wyatt, R.; Cayabyab, M.; Gerard, N.P.; Gerard, C.; Sodroski, J.; Choe, H. Tyrosine sulfation of the amino terminus of CCR5 facilitates HIV-1 entry. Cell 1999, 96, 667–676. [Google Scholar] [CrossRef]
- Liu, X.; Malins, L.R.; Roche, M.; Sterjovski, J.; Duncan, R.; Garcia, M.L.; Barnes, N.C.; Anderson, D.A.; Stone, M.J.; Gorry, P.R.; et al. Site-selective solid-phase synthesis of a CCR5 sulfopeptide library to interrogate HIV binding and entry. ACS Chem. Biol. 2014, 9, 2074–2081. [Google Scholar] [CrossRef] [PubMed]
- Farzan, M.; Chung, S.; Li, W.; Vasilieva, N.; Wright, P.L.; Schnitzler, C.E.; Marchione, R.J.; Gerard, C.; Gerard, N.P.; Sodroski, J.; et al. Tyrosine-sulfated peptides functionally reconstitute a CCR5 variant lacking a critical amino-terminal region. J. Biol. Chem. 2002, 277, 40397–40402. [Google Scholar] [CrossRef] [PubMed]
- Cormier, E.G.; Persuh, M.; Thompson, D.A.; Lin, S.W.; Sakmar, T.P.; Olson, W.C.; Dragic, T. Specific interaction of CCR5 amino-terminal domain peptides containing sulfotyrosines with HIV-1 envelope glycoprotein GP120. Proc. Natl. Acad. Sci. USA 2000, 97, 5762–5767. [Google Scholar] [CrossRef] [PubMed]
- Ju, T.; Niu, W.; Cerny, R.; Bollman, J.; Roy, A.; Guo, J. Molecular recognition of sulfotyrosine and phosphotyrosine by the SRC homology 2 domain. Mol. Biosyst. 2013, 9, 1829–1832. [Google Scholar] [CrossRef] [PubMed]
- Ju, T.; Niu, W.; Guo, J. Evolution of SRC homology 2 (SH2) domain to recognize sulfotyrosine. ACS Chem. Biol. 2016, 11, 2551–2557. [Google Scholar] [CrossRef] [PubMed]
- Rapp, C.; Klerman, H.; Levine, E.; McClendon, C.L. Hydrogen bond strengths in phosphorylated and sulfated amino acid residues. PLoS ONE 2013, 8. [Google Scholar] [CrossRef] [PubMed]
- Ludeman, J.P.; Nazari-Robati, M.; Wilkinson, B.L.; Huang, C.; Payne, R.J.; Stone, M.J. Phosphate modulates receptor sulfotyrosine recognition by the chemokine monocyte chemoattractant protein-1 (MCP-1/CCL2). Org. Biomol. Chem. 2015, 13, 2162–2169. [Google Scholar] [CrossRef] [PubMed]
- Crump, M.P.; Gong, J.H.; Loetscher, P.; Rajarathnam, K.; Amara, A.; Arenzana-Seisdedos, F.; Virelizier, J.L.; Baggiolini, M.; Sykes, B.D.; Clark-Lewis, I. Solution structure and basis for functional activity of stromal cell-derived factor-1; dissociation of CXCR4 activation from binding and inhibition of HIV-1. EMBO J. 1997, 16, 6996–7007. [Google Scholar] [CrossRef] [PubMed]
- Kleist, A.B.; Getschman, A.E.; Ziarek, J.J.; Nevins, A.M.; Gauthier, P.A.; Chevigne, A.; Szpakowska, M.; Volkman, B.F. New paradigms in chemokine receptor signal transduction: Moving beyond the two-site model. Biochem. Pharmacol. 2016, 114, 53–68. [Google Scholar] [CrossRef] [PubMed]
- Veldkamp, C.T.; Ziarek, J.J.; Peterson, F.C.; Chen, Y.; Volkman, B.F. Targeting SDF-1/CXCL12 with a ligand that prevents activation of CXCR4 through structure-based drug design. J. Am. Chem. Soc. 2010, 132, 7242–7243. [Google Scholar] [CrossRef] [PubMed]
- Smith, E.W.; Liu, Y.; Getschman, A.E.; Peterson, F.C.; Ziarek, J.J.; Li, R.; Volkman, B.F.; Chen, Y. Structural analysis of a novel small molecule ligand bound to the CXCL12 chemokine. J. Med. Chem. 2014, 57, 9693–9699. [Google Scholar] [CrossRef] [PubMed]
- Smith, E.W.; Nevins, A.M.; Qiao, Z.; Liu, Y.; Getschman, A.E.; Vankayala, S.L.; Kemp, M.T.; Peterson, F.C.; Li, R.; Volkman, B.F.; et al. Structure-based identification of novel ligands targeting multiple sites within a chemokine-G-protein-coupled-receptor interface. J. Med. Chem. 2016, 59, 4342–4351. [Google Scholar] [CrossRef] [PubMed]
- Smith, E.W.; Lewandowski, E.M.; Moussouras, N.A.; Kroeck, K.G.; Volkman, B.F.; Veldkamp, C.T.; Chen, Y. Crystallographic structure of truncated CCL21 and the putative sulfotyrosine-binding site. Biochemistry 2016, 55, 5746–5753. [Google Scholar] [CrossRef] [PubMed]
- Ziarek, J.J.; Kleist, A.B.; London, N.; Raveh, B.; Montpas, N.; Bonneterre, J.; St-Onge, G.; DiCosmo-Ponticello, C.J.; Koplinski, C.A.; Roy, I.; et al. Structural basis for chemokine recognition by a G protein-coupled receptor and implications for receptor activation. Sci. Signal. 2017, 10. [Google Scholar] [CrossRef] [PubMed]
- Ziarek, J.J.; Liu, Y.; Smith, E.; Zhang, G.; Peterson, F.C.; Chen, J.; Yu, Y.; Chen, Y.; Volkman, B.F.; Li, R. Fragment-based optimization of small molecule CXCL12 inhibitors for antagonizing the CXCL12/CXCR4 interaction. Curr. Top. Med. Chem. 2012, 12, 2727–2740. [Google Scholar] [CrossRef] [PubMed]
- Chang, W.C.; Lee, T.Y.; Shien, D.M.; Hsu, J.B.; Horng, J.T.; Hsu, P.C.; Wang, T.Y.; Huang, H.D.; Pan, R.L. Incorporating support vector machine for identifying protein tyrosine sulfation sites. J. Comput. Chem. 2009, 30, 2526–2537. [Google Scholar] [CrossRef] [PubMed]
- Monigatti, F.; Gasteiger, E.; Bairoch, A.; Jung, E. The sulfinator: Predicting tyrosine sulfation sites in protein sequences. Bioinformatics 2002, 18, 769–770. [Google Scholar] [CrossRef] [PubMed]
- Love, M.; Sandberg, J.L.; Ziarek, J.J.; Gerarden, K.P.; Rode, R.R.; Jensen, D.R.; McCaslin, D.R.; Peterson, F.C.; Veldkamp, C.T. Solution structure of CCL21 and identification of a putative CCR7 binding site. Biochemistry 2012, 51, 733–735. [Google Scholar] [CrossRef] [PubMed]
- Hauser, M.A.; Legler, D.F. Common and biased signaling pathways of the chemokine receptor CCR7 elicited by its ligands CCL19 and CCL21 in leukocytes. J. Leukoc. Biol. 2016, 99. [Google Scholar] [CrossRef] [PubMed]
- Hjorto, G.M.; Larsen, O.; Steen, A.; Daugvilaite, V.; Berg, C.; Fares, S.; Hansen, M.; Ali, S.; Rosenkilde, M.M. Differential CCR7 targeting in dendritic cells by three naturally occurring CC-chemokines. Front. Immunol. 2016, 7, 568. [Google Scholar] [CrossRef] [PubMed]
- Lorenz, N.; Loef, E.J.; Kelch, I.D.; Verdon, D.J.; Black, M.M.; Middleditch, M.J.; Greenwood, D.R.; Graham, E.S.; Brooks, A.E.; Dunbar, P.R.; et al. Plasmin and regulators of plasmin activity control the migratory capacity and adhesion of human T cells and dendritic cells by regulating cleavage of the chemokine CCL21. Immunol. Cell Biol. 2016, 94, 955–963. [Google Scholar] [CrossRef] [PubMed]
- Barmore, A.J.; Castex, S.M.; Gouletas, B.A.; Griffith, A.J.; Metz, S.W.; Muelder, N.G.; Populin, M.J.; Sackett, D.M.; Schuster, A.M.; Veldkamp, C.T. Transferring the C-terminus of the chemokine CCL21 to CCL19 confers enhanced heparin binding. Biochem. Biophys. Res. Commun. 2016, 477, 602–606. [Google Scholar] [CrossRef] [PubMed]
- Hauser, M.A.; Kindinger, I.; Laufer, J.M.; Spate, A.K.; Bucher, D.; Vanes, S.L.; Krueger, W.A.; Wittmann, V.; Legler, D.F. Distinct CCR7 glycosylation pattern shapes receptor signaling and endocytosis to modulate chemotactic responses. J. Leukoc. Biol. 2016, 99, 993–1007. [Google Scholar] [CrossRef] [PubMed]
- Kiermaier, E.; Moussion, C.; Veldkamp, C.T.; Gerardy-Schahn, R.; de Vries, I.; Williams, L.G.; Chaffee, G.R.; Phillips, A.J.; Freiberger, F.; Imre, R.; et al. Polysialylation controls dendritic cell trafficking by regulating chemokine recognition. Science 2016, 351, 186–190. [Google Scholar] [CrossRef] [PubMed]
- Veldkamp, C.T.; Koplinski, C.A.; Jensen, D.R.; Peterson, F.C.; Smits, K.M.; Smith, B.L.; Johnson, S.K.; Lettieri, C.; Buchholz, W.G.; Solheim, J.C.; et al. Production of recombinant chemokines and validation of refolding. Methods Enzymol. 2016, 570, 539–565. [Google Scholar] [PubMed]
- Birkenbach, M.; Josefsen, K.; Yalamanchili, R.; Lenoir, G.; Kieff, E. Epstein-barr virus-induced genes: First lymphocyte-specific G protein-coupled peptide receptors. J. Virol. 1993, 67, 2209–2220. [Google Scholar] [PubMed]
- Markley, J.L.; Ulrich, E.L.; Westler, W.M.; Volkman, B.F. Macromolecular structure determination by NMR spectroscopy. Methods Biochem. Anal. 2003, 44, 89–113. [Google Scholar] [PubMed]
- Veldkamp, C.T.; Peterson, F.C.; Pelzek, A.J.; Volkman, B.F. The monomer-dimer equilibrium of stromal cell-derived factor-1 (CXCL12) is altered by PH, phosphate, sulfate, and heparin. Protein Sci. 2005, 14, 1071–1081. [Google Scholar] [CrossRef] [PubMed]
- Veldkamp, C.T.; Kiermaier, E.; Gabel-Eissens, S.J.; Gillitzer, M.L.; Lippner, D.R.; DiSilvio, F.A.; Mueller, C.J.; Wantuch, P.L.; Chaffee, G.R.; Famiglietti, M.W.; et al. Solution structure of CCL19 and identification of overlapping CCR7 and PSGL-1 binding sites. Biochemistry 2015, 54, 4163–4166. [Google Scholar] [CrossRef] [PubMed]





| CCR7 | Kd (μM) 1 ± CI 2 |
|---|---|
| 5–11 | 700 ± 300 |
| 5–11 pY8 | 240 ± 60 |
| 5–11 sY8 | 140 ± 40 |
| 11–30 | 6,800 ± 500 |
| 11–30 pY17 | 1,700 ± 400 |
| 11–30 sY17 | 480 ± 70 |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Phillips, A.J.; Taleski, D.; Koplinski, C.A.; Getschman, A.E.; Moussouras, N.A.; Richard, A.M.; Peterson, F.C.; Dwinell, M.B.; Volkman, B.F.; Payne, R.J.; et al. CCR7 Sulfotyrosine Enhances CCL21 Binding. Int. J. Mol. Sci. 2017, 18, 1857. https://doi.org/10.3390/ijms18091857
Phillips AJ, Taleski D, Koplinski CA, Getschman AE, Moussouras NA, Richard AM, Peterson FC, Dwinell MB, Volkman BF, Payne RJ, et al. CCR7 Sulfotyrosine Enhances CCL21 Binding. International Journal of Molecular Sciences. 2017; 18(9):1857. https://doi.org/10.3390/ijms18091857
Chicago/Turabian StylePhillips, Andrew J., Deni Taleski, Chad A. Koplinski, Anthony E. Getschman, Natasha A. Moussouras, Amanda M. Richard, Francis C. Peterson, Michael B. Dwinell, Brian F. Volkman, Richard J. Payne, and et al. 2017. "CCR7 Sulfotyrosine Enhances CCL21 Binding" International Journal of Molecular Sciences 18, no. 9: 1857. https://doi.org/10.3390/ijms18091857
APA StylePhillips, A. J., Taleski, D., Koplinski, C. A., Getschman, A. E., Moussouras, N. A., Richard, A. M., Peterson, F. C., Dwinell, M. B., Volkman, B. F., Payne, R. J., & Veldkamp, C. T. (2017). CCR7 Sulfotyrosine Enhances CCL21 Binding. International Journal of Molecular Sciences, 18(9), 1857. https://doi.org/10.3390/ijms18091857