The Future of DNA Adductomic Analysis
Abstract
:1. Introduction
2. Conventional Approach for DNA Adduct Screening: 32P-Postlabeling
3. New Approach for DNA Adduct Screening: DNA Adductomics Using Liquid Chromatography-Mass Spectrometry (LC-MS)
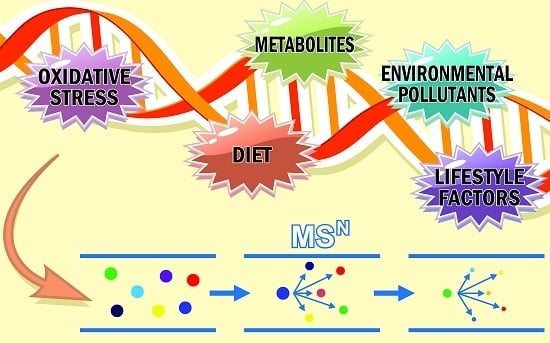
3.1. Typical DNA Adductomics Workflow
3.2. Key Feature of the Positive Ion LC-MSn DNA Adductomics Methodology
3.3. A Sensitive and Selective LC-MSn Screening
3.4. Rapidly Evolving Technology
3.4.1. Nanospray Ionization
3.4.2. High Resolution Accurate Mass (HRAM) Data
3.4.3. Scanning Modes for HRAM MSn Data Acquisition
3.4.4. Data Dependent Acquisition (MSn)
3.4.5. Data Independent Acquisition (MS2)
3.4.6. DDA and DIA for DNA Adductomics
4. Adductomic Studies
Need for Methodology Comparisons
5. New HRAM DNA Adductomic Studies
5.1. Untargeted and Targeted Nanospray HRAM CNL-MS3 Analysis
5.2. Untargeted HRAM MSE Analysis
5.3. Targeted HRAM Full Scan Analysis
5.4. Adduct-Tagging MALDI Ionization Approach
6. Challenges
6.1. Selectivity
6.2. Sensitivity
6.3. Quantitation
6.4. Ease of Data Analysis
7. Summary
Acknowledgments
Conflicts of Interest
Abbreviations
| LC-MS | Liquid chromatography—mass spectrometry |
| LC-MS2 | Liquid chromatography—tandem mass spectrometry |
| LC-MSn | Liquid chromatography—multistage fragmentation mass spectrometry |
| HRAM | High resolution/accurate mass |
| MALDI | Matrix assisted laser desorption ionization |
| TOF | Time-of-flight |
| UPLC | Ultra high pressure liquid chromatography |
| Q-TOF | Quadrupole-Time-of-flight |
| DDA | Data dependent acquisition |
| DIA | Data independent acquisition |
| CNL/MS3 | Constant neutral loss/triple stage mass spectrometry |
| SIM/MS2 | Selected ion monitoring/tandem mass spectrometry |
| NNK | Nitrosamine 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone |
| LPS | Lipopolysaccharide |
| PCA | Principal component analysis |
| MALDI-TOF | Matrix assisted laser desorption ionization—time-of-flight |
| MALDI-TOF/TOF | Matrix assisted laser desorption ionization—time-of-flight/time-of-flight |
References
- Wiencke, J.K. DNA adduct burden and tobacco carcinogenesis. Oncogene 2002, 21, 7376–7391. [Google Scholar] [CrossRef] [PubMed]
- Dipple, A. DNA adduts of chemical carcinogens. Carcinogenesis 1995, 16, 437–441. [Google Scholar] [CrossRef] [PubMed]
- Poirier, M.C. Linking DNA adduct formation and human cancer risk in chemical carcinogenesis. Environ. Mol. Mutagen. 2016, 57, 499–507. [Google Scholar] [CrossRef] [PubMed]
- Poirier, M.C.; Beland, F.A. DNA adduct measurements and tumor incidence during chronic carcinogen exposure in rodents. Environ. Health. Perspect. 1994, 102 (Suppl. S6), 161–165. [Google Scholar] [CrossRef] [PubMed]
- Hemminki, K.; Koskinen, M.; Rajaniemi, H.; Zhao, C. DNA adducts, mutations, and cancer 2000. Regul. Toxicol. Pharmacol. 2000, 32, 264–275. [Google Scholar] [CrossRef] [PubMed]
- Food and Drug Administration, HHS. International conference on harmonisation; guidance on s2(r1) genotoxicity testing and data interpretation for pharmaceuticals intended for human use; availability. Notice. Fed. Regist. 2012, 77, 33748–33749. [Google Scholar]
- Klaene, J.J.; Sharma, V.K.; Glick, J.; Vouros, P. The analysis of DNA adducts: The transition from 32P-postlabeling to mass spectrometry. Cancer Lett. 2013, 334, 10–19. [Google Scholar] [CrossRef] [PubMed]
- Farmer, P.B.; Singh, R. Use of DNA adducts to identify human health risk from exposure to hazardous environmental pollutants: The increasing role of mass spectrometry in assessing biologically effective doses of genotoxic carcinogens. Mutat. Res. 2008, 659, 68–76. [Google Scholar] [CrossRef] [PubMed]
- Parry, J.M. Genetic Toxicol. Principles Methods; Humana: Totowa, NJ, USA, 2012; p. 433. [Google Scholar]
- Rybicki, B.A.; Rundle, A.; Savera, A.T.; Sankey, S.S.; Tang, D. Polycyclic aromatic hydrocarbon-DNA adducts in prostate cancer. Cancer Res. 2004, 64, 8854–8859. [Google Scholar] [CrossRef] [PubMed]
- Blommaert, F.A.; Michael, C.; Terheggen, P.M.; Muggia, F.M.; Kortes, V.; Schornagel, J.H.; Hart, A.A.; den Engelse, L. Drug-induced DNA modification in buccal cells of cancer patients receiving carboplatin and cisplatin combination chemotherapy, as determined by an immunocytochemical method: Interindividual variation and correlation with disease response. Cancer Res. 1993, 53, 5669–5675. [Google Scholar] [PubMed]
- Andrews, C.L.; Vouros, P.; Harsch, A. Analysis of DNA adducts using high-performance separation techniques coupled to electrospray ionization mass spectrometry. J. Chromatogr. 1999, 856, 515–526. [Google Scholar] [CrossRef]
- Koc, H.; Swenberg, J.A. Applications of mass spectrometry for quantitation of DNA adducts. J. Chromatogr. B 2002, 778, 323–343. [Google Scholar] [CrossRef]
- Singh, R.; Farmer, P.B. Liquid chromatography-electrospray ionization-mass spectrometry: The future of DNA adduct detection. Carcinogenesis 2006, 27, 178–196. [Google Scholar] [CrossRef] [PubMed]
- Tretyakova, N.; Goggin, M.; Sangaraju, D.; Janis, G. Quantitation of DNA adducts by stable isotope dilution mass spectrometry. Chem. Res. Toxicol. 2012, 25, 2007–2035. [Google Scholar] [CrossRef] [PubMed]
- Tretyakova, N.; Villalta, P.W.; Kotapati, S. Mass spectrometry of structurally modified DNA. Chem. Rev. 2013, 113, 2395–2436. [Google Scholar] [CrossRef] [PubMed]
- Beach, A.C.; Gupta, R.C. Human biomonitoring and the 32P-postlabeling assay. Carcinogenesis 1992, 13, 1053–1074. [Google Scholar] [CrossRef] [PubMed]
- Phillips, D.H. DNA adducts as markers of exposure and risk. Mutat. Res. 2005, 577, 284–292. [Google Scholar] [CrossRef] [PubMed]
- Jones, N.J.; McGregor, A.D.; Waters, R. Detection of DNA adducts in human oral tissue: Correlation of adduct levels with tobacco smoking and differential enhancement of adducts using the butanol extraction and nuclease p1 versons of 32p postlabeling. Cancer Res. 1993, 53, 1522–1528. [Google Scholar] [PubMed]
- Phillips, D.H. Smoking-related DNA and protein adducts in human tissues. Carcinogenesis 2002, 23, 1979–2004. [Google Scholar] [CrossRef] [PubMed]
- Di Paolo, O.A.; Teitel, C.H.; Nowell, S.; Coles, B.F.; Kadlubar, F.F. Expression of cytochromes P450 and glutathione s-transferases in human prostate, and the potential for activation of heterocyclic amine carcinogens via acetyl-coa-, paps- and atp-dependent pathways. Int. J. Cancer 2005, 117, 8–13. [Google Scholar] [CrossRef] [PubMed]
- Nath, R.G.; Ocando, J.E.; Guttenplan, J.B.; Chung, F.L. 1,n2-propanodeoxyguanosine adducts: Potential new biomarkers of smoking-induced DNA damage in human oral tissue. Cancer Res. 1998, 58, 581–584. [Google Scholar] [PubMed]
- Arif, J.M.; Dresler, C.; Clapper, M.L.; Gairola, C.G.; Srinivasan, C.; Lubet, R.A.; Gupta, R.C. Lung DNA adducts detected in human smokers are unrelated to typical polyaromatic carcinogens. Chem. Res. Toxicol. 2006, 19, 295–299. [Google Scholar] [CrossRef] [PubMed]
- Talaska, G.; Schamer, M.; Skipper, P.; Tannenbaum, S.; Caporaso, N.; Unruh, L.; Kadlubar, F.F.; Bartsch, H.; Malaveille, C.; Vineis, P. Detection of carcinogen-DNA adducts in exfoliated urothelial cells of cigarette smokers: Association with smoking, hemoglobin adducts, and urinary mutagenicity. Cancer Epidemiol. Biomark. Prev. 1991, 1, 61–66. [Google Scholar] [PubMed]
- Gorlewska-Roberts, K.; Green, B.; Fares, M.; Ambrosone, C.B.; Kadlubar, F.F. Carcinogen-DNA adducts in human breast epithelial cells. Environ. Mol. Mutagen. 2002, 39, 184–192. [Google Scholar] [CrossRef] [PubMed]
- Nia, A.B.; Maas, L.M.; van Breda, S.G.; Curfs, D.M.; Kleinjans, J.C.; Wouters, E.F.; van Schooten, F.J. Applicability of induced sputum for molecular dosimetry of exposure to inhalatory carcinogens: 32P-postlabeling of lipophilic DNA adducts in smokers and nonsmokers. Cancer Epidemiol. Biomark. Prev. 2000, 9, 367–372. [Google Scholar]
- Nia, A.B.; Maas, L.M.; Brouwer, E.M.; Kleinjans, J.C.; van Schooten, F.J. Comparison between smoking-related DNA adduct analysis in induced sputum and peripheral blood lymphocytes. Carcinogenesis 2000, 21, 1335–1340. [Google Scholar] [CrossRef]
- Phillips, D.H. On the origins and development of the 32p-postlabelling assay for carcinogen-DNA adducts. Cancer Lett. 2013, 334, 5–9. [Google Scholar] [CrossRef] [PubMed]
- Balbo, S.; Turesky, R.J.; Villalta, P.W. DNA adductomics. Chem. Res. Toxicol. 2014, 27, 356–366. [Google Scholar] [CrossRef] [PubMed]
- Totsuka, Y.; Fukutome, K.; Takahashi, M.; Takahashi, S.; Tada, A.; Sugimura, T.; Wakabayashi, K. Presence of N2-(deoxyguanosin-8-yl)-2-amino-3,8-dimethylimidazo[4,5-f]quinoxaline (dG-C8-MeIQx) in human tissues. Carcinogenesis 1996, 17, 1029–1034. [Google Scholar] [CrossRef] [PubMed]
- Phillips, D.H.; Arlt, V.M. The 32P-postlabeling assay for DNA adducts. Nat. Protocols 2007, 2, 2772–2781. [Google Scholar] [CrossRef] [PubMed]
- Claereboudt, J.; Esmans, E.L.; Vandeneeckhout, E.G.; Claeys, M. Fast-atom-bombardment and tandem mass-spectrometry for the identification of nucleoside adducts with phenyl glycidyl ether. Nucleos. Nucleot. 1990, 9, 333–344. [Google Scholar] [CrossRef]
- Hemeryck, L.Y.; Rombouts, C.; van Hecke, T.; van Meulebroek, L.; Vanden Bussche, J.; de Smet, S.; Vanhaecke, L. In vitro DNA adduct profiling to mechanistically link red meat consumption to colon cancer promotion. Toxicol. Res. 2016, 5, 1346–1358. [Google Scholar] [CrossRef]
- Hemeryck, L.Y.; Decloedt, A.I.; Vanden Bussche, J.; Geboes, K.P.; Vanhaecke, L. High resolution mass spectrometry based profiling of diet-related deoxyribonucleic acid adducts. Anal. Chim. Acta 2015, 892, 123–131. [Google Scholar] [CrossRef] [PubMed]
- Ishino, K.; Kato, T.; Kato, M.; Shibata, T.; Watanabe, M.; Wakabayashi, K.; Nakagama, H.; Totsuka, Y. Comprehensive DNA adduct analysis reveals pulmonary inflammatory response contributes to genotoxic action of magnetite nanoparticles. Int. J. Mol. Sci. 2015, 16, 3474–3492. [Google Scholar] [CrossRef] [PubMed]
- Balbo, S.; Hecht, S.S.; Upadhyaya, P.; Villalta, P.W. Application of a high-resolution mass-spectrometry-based DNA adductomics approach for identification of DNA adducts in complex mixtures. Anal. Chem. 2014, 86, 1744–1752. [Google Scholar] [CrossRef] [PubMed]
- Stornetta, A.; Villalta, P.W.; Hecht, S.S.; Sturla, S.; Balbo, S. Screening for DNA alkylation mono and cross-linked adducts with a comprehensive LC-MS3 adductomic approach. Anal. Chem. 2015, 87, 11706–11713. [Google Scholar] [CrossRef] [PubMed]
- Carra’, A.; Villalta, P.W.; Dator, R.P.; Balbo, S. Screening for inflammation-induced DNA adducts with a comprehensive high resolution LC-MS3 adductomic approach. In Proceedings of the 64th ASMS Annual Meeting, San Antonio, TX, USA, 4–8 June 2016. [Google Scholar]
- Gates, K.S.; Nooner, T.; Dutta, S. Biologically relevant chemical reactions of n7-alkylguanine residues in DNA. Chem. Res. Toxicol 2004, 17, 839–856. [Google Scholar] [CrossRef] [PubMed]
- Inagaki, S.; Hirashima, H.; Esaka, Y.; Higashi, T.; Min, J.Z.; Toyo‘oka, T. Screening DNA adducts by lc–esi–ms–ms: Application to screening new adducts formed from acrylamide. Chromatographia 2010, 72, 1043–1048. [Google Scholar] [CrossRef]
- Gangl, E.T.; Turesky, R.J.; Vouros, P. Determination of in vitro- and in vivo-formed DNA adducts of 2-amino-3-methylimidazo[4,5-f]quinoline by Capillary Liquid Chromatography/Microelectrospray Mass Spectrometry. Chem. Res. Toxicol. 1999, 12, 1019–1027. [Google Scholar] [CrossRef] [PubMed]
- Shi, T.; Song, E.; Nie, S.; Rodland, K.D.; Liu, T.; Qian, W.J.; Smith, R.D. Advances in targeted proteomics and applications to biomedical research. Proteomics 2016, 16, 2160–2182. [Google Scholar] [CrossRef] [PubMed]
- Gillet, L.C.; Navarro, P.; Tate, S.; Rost, H.; Selevsek, N.; Reiter, L.; Bonner, R.; Aebersold, R. Targeted data extraction of the MS/MS spectra generated by data-independent acquisition: A new concept for consistent and accurate proteome analysis. Mol. Cell Proteom. 2012, 11, O111-016717. [Google Scholar] [CrossRef] [PubMed]
- Gillet, L.C.; Leitner, A.; Aebersold, R. Mass spectrometry applied to bottom-up proteomics: Entering the high-throughput era for hypothesis testing. Annu Rev. Anal. Chem. 2016, 9, 449–472. [Google Scholar] [CrossRef] [PubMed]
- Venable, J.D.; Dong, M.Q.; Wohlschlegel, J.; Dillin, A.; Yates, J.R. Automated approach for quantitative analysis of complex peptide mixtures from tandem mass spectra. Nat. Methods 2004, 1, 39–45. [Google Scholar] [CrossRef] [PubMed]
- Xiao, J.F.; Zhou, B.; Ressom, H.W. Metabolite identification and quantitation in LC-MS/MS-based metabolomics. Trends Anal. Chem. 2012, 32, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Tsugawa, H.; Cajka, T.; Kind, T.; Ma, Y.; Higgins, B.; Ikeda, K.; Kanazawa, M.; VanderGheynst, J.; Fiehn, O.; Arita, M. MS-DIAL: Data-independent MS/MS deconvolution for comprehensive metabolome analysis. Nat. Methods 2015, 12, 523–526. [Google Scholar] [CrossRef] [PubMed]
- Tsou, C.C.; Avtonomov, D.; Larsen, B.; Tucholska, M.; Choi, H.; Gingras, A.C.; Nesvizhskii, A.I. DIA-Umpire: comprehensive computational framework for data-independent acquisition proteomics. Nat. Methods 2015, 12, 258–264. [Google Scholar] [CrossRef] [PubMed]
- Bilbao, A.; Varesio, E.; Luban, J.; Strambio-De-Castillia, C.; Hopfgartner, G.; Muller, M.; Lisacek, F. Processing strategies and software solutions for data-independent acquisition in mass spectrometry. Proteomics 2015, 15, 964–980. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Cai, Y.; Guo, Y.; Chen, F.; Zhu, Z.J. MetDIA: Targeted metabolite extraction of multiplexed MS/MS spectra generated by data-independent acquisition. Anal. Chem. 2016, 88, 8757–8764. [Google Scholar] [CrossRef] [PubMed]
- Hemeryck, L.Y.; Moore, S.A.; Vanhaecke, L. Mass spectrometric mapping of the DNA adductome as a means to study genotoxin exposure, metabolism and effect. Anal. Chem. 2016, 88, 7436–7446. [Google Scholar] [CrossRef] [PubMed]
- Hemeryck, L.Y.; Vanhaecke, L. Diet-related DNA adduct formation in relation to carcinogenesis. Nutr. Rev. 2016, 74, 15. [Google Scholar] [CrossRef] [PubMed]
- Yao, C.; Feng, Y.-L. A nontargeted screening method for covalent DNA adducts and DNA modification selectivity using liquid chromatography-tandem mass spectrometry. Talanta 2016, 159, 10. [Google Scholar] [CrossRef] [PubMed]
- Yao, C.; Foster, W.G.; Sadeu, J.C.; Siddique, S.; Zhu, J.; Feng, Y.L. Screening for DNA adducts in ovarian follicles exposed to benzo [a] pyrene and cigarette smoke condensate using liquid chromatography-tandem mass spectrometry. Sci. Total Environ. 2016, 575, 742–749. [Google Scholar] [CrossRef] [PubMed]
- Kanaly, R.A.; Micheletto, R.; Matsuda, T.; Utsuno, Y.; Ozeki, Y.; Hamamura, N. Application of DNA adductomics to soil bacterium Sphingobium sp strain KK22. Microbiologyopen 2015, 4, 841–856. [Google Scholar] [CrossRef] [PubMed]
- Bessette, E.E.; Goodenough, A.K.; Langouet, S.; Yasa, I.; Kozekov, I.D.; Spivack, S.D.; Turesky, R.J. Screening for DNA adducts by data-dependent constant neutral loss-triple stage mass spectrometry with a linear quadrupole ion trap mass spectrometer. Anal. Chem. 2009, 81, 809–819. [Google Scholar] [CrossRef] [PubMed]
- Guo, J.; Villalta, P.W.; Turesky, R.J. A data-independent mass spectrometry approach for screening and identification of DNA adducts. Anal. Chem. submitted.
- Bryant, M.S.; Lay, J.O.; Chiarelli, M.P. Development of fast atom bombardment mass spectral methods for the identification of carcinogen-nucleoside adducts. J. Am. Soc. Mass Spectrom. 1992, 3, 360–371. [Google Scholar] [CrossRef]
- Rindgen, D.; Turesky, R.J.; Vouros, P. Determination of in vitro formed DNA adducts of 2-amino-1-methyl-6-phenylimidazo [4,5-b] pyridine using capillary liquid chromatography/electrospray ionization/tandem mass spectrometry. Chem. Res. Toxicol. 1995, 8, 1005–1013. [Google Scholar] [CrossRef] [PubMed]
- Regulus, P.; Spessotto, S.; Gateau, M.; Cadet, J.; Favier, A.; Ravanat, J.L. Detection of new radiation-induced DNA lesions by liquid chromatography coupled to tandem mass spectrometry. Rapid Commun. Mass Spectrom. 2004, 18, 2223–2228. [Google Scholar] [CrossRef] [PubMed]
- Singh, R.; Teichert, F.; Seidel, A.; Roach, J.; Cordell, R.; Cheng, M.K.; Frank, H.; Steward, W.P.; Manson, M.M.; Farmer, P.B. Development of a targeted adductomic method for the determination of polycyclic aromatic hydrocarbon DNA adducts using online column-switching liquid chromatography/tandem mass spectrometry. Rapid Commun. Mass Spectrom. 2010, 24, 2329–2340. [Google Scholar] [CrossRef] [PubMed]
- Kanaly, R.A.; Hanaoka, T.; Sugimura, H.; Toda, H.; Matsui, S.; Matsuda, T. Development of the adductome approach to detect DNA damage in humans. Antioxid. Redox. Signal 2006, 8, 993–1001. [Google Scholar] [CrossRef] [PubMed]
- Kanaly, R.A.; Matsui, S.; Hanaoka, T.; Matsuda, T. Application of the adductome approach to assess intertissue DNA damage variations in human lung and esophagus. Mutat. Res. 2007, 625, 83–93. [Google Scholar] [CrossRef] [PubMed]
- Chou, P.H.; Kageyama, S.; Matsuda, S.; Kanemoto, K.; Sasada, Y.; Oka, M.; Shinmura, K.; Mori, H.; Kawai, K.; Kasai, H.; et al. Detection of lipid peroxidation-induced DNA adducts caused by 4-oxo-2 (E)-nonenal and 4-oxo-2 (E)-hexenal in human autopsy tissues. Chem. Res. Toxicol. 2010, 23, 1442–1448. [Google Scholar] [CrossRef] [PubMed]
- Spilsberg, B.; Rundberget, T.; Johannessen, L.E.; Kristoffersen, A.B.; Holst-Jensen, A.; Berdal, K.G. Detection of food-derived damaged nucleosides with possible adverse effects on human health using a global adductomics approach. J. Agric. Food Chem. 2010, 58, 6370–6375. [Google Scholar] [CrossRef] [PubMed]
- Kato, K.; Yamamura, E.; Kawanishi, M.; Yagi, T.; Matsuda, T.; Sugiyama, A.; Uno, Y. Application of the DNA adductome approach to assess the DNA-damaging capability of in vitro micronucleus test-positive compounds. Mutat. Res. 2011, 721, 21–26. [Google Scholar] [CrossRef] [PubMed]
- Matsuda, T.; Tao, H.; Goto, M.; Yamada, H.; Suzuki, M.; Wu, Y.; Xiao, N.; He, Q.; Guo, W.; Cai, Z.; et al. Lipid peroxidation-induced DNA adducts in human gastric mucosa. Carcinogenesis 2013, 34, 121–127. [Google Scholar] [CrossRef] [PubMed]
- Van den Driessche, B.; van Dongen, W.; Lemiere, F.; Esmans, E.L. Implementation of data-dependent acquisitions in the study of melphalan DNA adducts by miniaturized liquid chromatography coupled to electrospray tandem mass spectrometry. Rapid Commun. Mass Spectrom. 2004, 18, 2001–2007. [Google Scholar] [CrossRef] [PubMed]
- Pietsch, K.E.; van Midwoud, P.M.; Villalta, P.W.; Sturla, S.J. Quantification of acylfulvene- and illudin s-DNA adducts in cells with variable bioactivation capacities. Chem. Res. Toxicol. 2013, 26, 146–155. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.; Gao, J.; Li, G.; Shimelis, O.; Giese, R.W. Nontargeted analysis of DNA adducts by mass-tag ms: Reaction of p-benzoquinone with DNA. Chem. Res. Toxicol. 2012, 25, 2737–2743. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.; Fisher, D.; Rao, A.; Giese, R.W. Nontargeted nucleotide analysis based on benzoylhistamine labeling-MALDI-TOF/TOF-MS: discovery of putative 6-oxo-thymine in DNA. Anal. Chem. 2012, 84, 3811–3819. [Google Scholar] [CrossRef] [PubMed]
- Klaene, J.J.; Flarakos, C.; Glick, J.; Barret, J.T.; Zarbl, H.; Vouros, P. Tracking matrix effects in the analysis of DNA adducts of polycyclic aromatic hydrocarbons. J. Chromatogr. A 2016, 1439, 112–123. [Google Scholar] [CrossRef] [PubMed]
- Villalta, P.W.H.; Hochalter, J.B.; Hecht, S.S. Ultra-sensitive high resolution mass spectrometric analysis of a DNA adduct of the carcinogen benzo [a] pyrene in human lung. Anal. Chem. submitted.
- Stornetta, A.; Villalta, P.W.; Gossner, F.; Wilson, W.R.; Balbo, S.; Sturla, S.J. DNA adduct profiles predict in vitro cell viability after treatment with the experimental anticancer prodrug pr104a. Chem. Res. Toxicol. 2017, 30, 830–839. [Google Scholar] [CrossRef] [PubMed]




| Approach | Method | Scan Events | Frequency | Adduct Detection |
|---|---|---|---|---|
| DDA CNL/MS3 | Targeted | Full Scan | Continuous | MS/MS/MS Triggered Event |
| MS/MS | Ions included in a list | |||
| MS/MS/MS | MS/MS ions selected by loss of 116.0474 | |||
| Untargeted | Full Scan | Continuous | MS/MS/MS Triggered Event | |
| MS/MS | Most abundant ions | |||
| MS/MS/MS | MS/MS ions selected by loss of 116.0473 | |||
| DIA Wide SIM/MS2 | Targeted | Full Scan | Continuous | Post-run data analysis on ions from a list (characterized by co-eluters with NL = 116.0473) |
| MS/MS | Continuous | |||
| Untargeted | Full Scan | Continuous | Post-run data analysis (any co-eluters with NL = 116.0473) | |
| MS/MS | Continuous |
| Approach | Instrument | Sample Type | Adduct Type/Origin | Strengths | Weaknesses | Details | Reference |
|---|---|---|---|---|---|---|---|
| CNL | DF-EB/Q | Reaction with nucleosides | PGE c (industrial chemical) | High resolution, First example of DNA adductomic analysis | Simplistic model | Nucleoside reacted with chemical of interest | Claereboudt et al., 1990 [32] |
| Triple Quad | Synthetic standards | Arylamine (industrial chemical) | Early report of DNA adductomics | Nominal mass measurement and lack of fragmentation data | Analysis of synthetic standards only | Bryant et al., 1992 [58] | |
| In vitro reaction | PhIP a (food) | Comparison made with 32P-postlabeling | - | Vouros et al., 1995 [59] | |||
| In vitro reaction and Animal tissues | IQ b (food) | First example of nanospray ionization | - | Vouros et al., 1999 [41] | |||
| Irradiated cells (human monocyte) | Radiation-induced | Only example of analysis of adducts due to exposure to radiation | - | Ravanat et al., 2004 [60] | |||
| In vitro reaction | PAH (environmental/industrial exposure) | Automated data analysis | Small mass range (500–650 Da) | Singh et al., 2010 [61] | |||
| Reaction with oligonucleotide | PGE c, SO d (industrial chemicals) | - | Limited to oligonucleotides | Feng et al., 2016 [53] | |||
| Treated cells (from ovarian follicles) | PAH e (environmental/industrial exposure) | - | - | Feng et al., 2016 [54] | |||
| Pseudo-CNL | Human lung tissue | Screening for all DNA modifications | Adductome map data analysis | - | Matsuda et al., 2006 [62] | ||
| Human lung and esophagus tissue | Screening for all DNA modifications | Seven adducts unambiguously detected | - | Matsuda et al., 2007 [63] | |||
| Various human tissues | LPO-induced (endogenous) | Reported lipid peroxidation-derived adducts in humans | - | Matsuda et al., 2010 [64] | |||
| Quorn, button mushrooms, brewer’s yeast | Food | - | Only 7 SRM transitions per injection | Berdal et al., 2010 [65] | |||
| Treated cells (Chinese hamster) | Micronucleus test-positive compounds | First comparison to micronucleus test | - | Yagi et al., 2011 [66] | |||
| Human gastric mucosa | LPO (endogenous) | - | - | Matsuda et al., 2013 [67] | |||
| Soil Bacterium | Screening for all DNA modifications | First DNA adductomic study of bacterial DNA | - | Kanaly et al., 2015 [55] | |||
| DD-MS2 | Q-TOF | Treated cells (immortalized human T lymphocyte) | Melphalan (chemotherapy drug) | First example of MS2 spectral data acquisition | No MS3 fragmentation data, accurate mass data not reported | - | Esmans et al., 2004 [68] |
| MSE (HRAM) | Mouse lung tissue | Magnetic nanoparticles | First application of MSE | No MS2 or MS3 data, reported accurate mass data limited to 10 mmu | - | Totsuka et al., 2015 [35] | |
| Full Scan (HRAM) | Orbitrap | Human colon tumor tissue | Diet-related | - | - | Diet-related DNA adduct database, acid hydrolysis resulting in nucleobase adducts | Vanhaecke et al., 2015 [34] |
| In vitro microbiota meat digests | Diet-related | - | - | Utilized methodology developed in [34] | Vanhaecke et al., 21016 [33] | ||
| DD-CNL-MS3 | Ion Trap | Treated cells (human hepatocytes) Rat liver Human buccal cells | 4-ABP f, MeIQx g Tobacco constituents | Human samples examined, First example of MS3 data acquisition | No accurate mass measurements | - | Turesky et al., 2009 [56] |
| Treated cells (human colon adenocarcinoma) | Illudin S (chemotherapeutic natural product) | - | Used similar method to Turesky [56] | Sturla et al., 2013 [69] | |||
| DD-CNL-MS3 (HRAM) | Orbitrap | Mouse liver tissue | Tobacco constituents | Combination of HRAM, MS3 and nanospray | Extensive sample purification and multiple injections | - | Balbo et al., 2014 [36] |
| Treated cells (human colon adenocarcinoma) | DNA alkylating drug | First targeted approach | - | - | Balbo et al., 2015 [37] | ||
| Mouse lung tissue | Endogenous adducts | HRAM MS3 data acquisition | - | - | Balbo et al., 2017 [38] |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Villalta, P.W.; Balbo, S. The Future of DNA Adductomic Analysis. Int. J. Mol. Sci. 2017, 18, 1870. https://doi.org/10.3390/ijms18091870
Villalta PW, Balbo S. The Future of DNA Adductomic Analysis. International Journal of Molecular Sciences. 2017; 18(9):1870. https://doi.org/10.3390/ijms18091870
Chicago/Turabian StyleVillalta, Peter W., and Silvia Balbo. 2017. "The Future of DNA Adductomic Analysis" International Journal of Molecular Sciences 18, no. 9: 1870. https://doi.org/10.3390/ijms18091870
APA StyleVillalta, P. W., & Balbo, S. (2017). The Future of DNA Adductomic Analysis. International Journal of Molecular Sciences, 18(9), 1870. https://doi.org/10.3390/ijms18091870