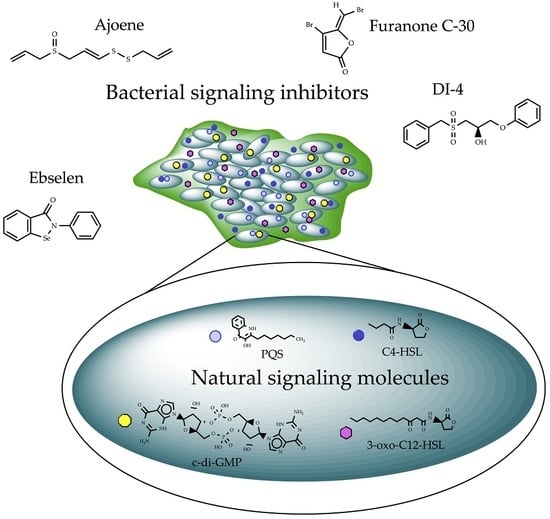
Bacterial Biofilm Control by Perturbation of Bacterial Signaling Processes
Abstract
:1. Introduction
2. Bacterial Biofilms
The Protective Biofilm Matrix of P. aeruginosa
3. Cellular Regulatory Systems Involved in the Biofilm Life-Cycle
3.1. Quorum Sensing and Its Role in the P. aeruginosa Biofilm
3.2. Bis-(3′-5′)-cyclic-dimeric Guanosine Monophosphate (Cyclic-di-GMP) and Its Role in P. aeruginosa Biofilm
3.3. Two-Component Systems and Small RNAs
3.4. Interconnections between the Regulatory Systems
4. Signal Perturbation by Small Molecules
4.1. Compounds Modulating Quorum Sensing (QS) Signaling
4.2. Compounds Targeting sRNAs
4.3. Compounds Modulating c-di-GMP Signaling
5. Potential Treatment Strategies by Cellular Signaling Perturbation
6. Final Remarks
Acknowledgments
Conflicts of Interest
References
- Potera, C. Studying slime. Environ. Health Perspect. 1998, 106, A604–A606. [Google Scholar] [CrossRef] [PubMed]
- Costerton, J.W.; Lewandowski, Z. Microbial biofilms. Annu. Rev. Microbiol. 1995, 49, 711–745. [Google Scholar] [CrossRef] [PubMed]
- Høiby, N.; Bjarnsholt, T.; Givskov, M.; Molin, S.; Ciofu, O. Antibiotic resistance of bacterial biofilms. Int. J. Antimicrob. Agents 2010, 35, 322–332. [Google Scholar] [CrossRef] [PubMed]
- Bjarnsholt, T.; Givskov, M. The role of quorum sensing in the pathogenicity of the cunning aggressor Pseudomonas aeruginosa. Anal. Bioanal. Chem. 2007, 387, 409–414. [Google Scholar] [CrossRef] [PubMed]
- Costerton, J.W.; Stewart, P.S.; Greenberg, E.P. Bacterial biofilms: A common cause of persistent infections. Science 1999, 284, 1318–1322. [Google Scholar] [CrossRef] [PubMed]
- Post, J.C. Direct evidence of bacterial biofilms in otitis media. Laryngoscope 2001, 111, 2083–2094. [Google Scholar] [CrossRef] [PubMed]
- Barbosa, F.C.; Mayer, M.P.; Saba-Chujfi, E.; Cai, S. Subgingival occurrence and antimicrobial susceptibility of enteric rods and pseudomonads from Brazilian periodontitis patients. Oral Microbiol. Immunol. 2001, 16, 306–310. [Google Scholar] [CrossRef] [PubMed]
- Lyczak, J.B.; Cannon, C.L.; Pier, G.B. Establishment of Pseudomonas aeruginosa infection: Lessons from a versatile opportunist. Microbes Infect. 2000, 2, 1051–1060. [Google Scholar] [CrossRef]
- Obritsch, M.D.; Fish, D.N.; MacLaren, R.; Jung, R. Nosocomial infections due to multidrug-resistant Pseudomonas aeruginosa: Epidemiology and treatment options. Pharmacotherapy 2005, 25, 1353–1364. [Google Scholar] [CrossRef] [PubMed]
- Woodward, T.C.; Brown, R.; Sacco, P.; Zhang, J. Budget impact model of tobramycin inhalation solution for treatment of Pseudomonas aeruginosa in cystic fibrosis patients. J. Med. Econ. 2010, 13, 492–499. [Google Scholar] [CrossRef] [PubMed]
- Koch, C.; Hoiby, N. Pathogenesis of cystic fibrosis. Lancet 1993, 341, 1065–1069. [Google Scholar] [CrossRef]
- Hentzer, M.; Wu, H.; Andersen, J.B.; Riedel, K.; Rasmussen, T.B.; Bagge, N.; Kumar, N.; Schembri, M.A.; Song, Z.; Kristoffersen, P.; et al. Attenuation of Pseudomonas aeruginosa virulence by quorum sensing inhibitors. EMBO J. 2003, 22, 3803–3815. [Google Scholar] [CrossRef] [PubMed]
- Hentzer, M.; Riedel, K.; Rasmussen, T.B.; Heydorn, A.; Andersen, J.B.; Parsek, M.R.; Rice, S.A.; Eberl, L.; Molin, S.; Høiby, N.; et al. Inhibition of quorum sensing in Pseudomonas aeruginosa biofilm bacteria by a halogenated furanone compound. Microbiology 2002, 148, 87–102. [Google Scholar] [CrossRef] [PubMed]
- Hentzer, M.; Givskov, M. Pharmacological inhibition of quorum sensing for the treatment of chronic bacterial infections. J. Clin. Investig. 2003, 112, 1300–1307. [Google Scholar] [CrossRef] [PubMed]
- Zuker, M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003, 31, 3406–3415. [Google Scholar] [CrossRef] [PubMed]
- Gómez, M.I.; Prince, A. Opportunistic infections in lung disease: Pseudomonas infections in cystic fibrosis. Curr. Opin. Pharmacol. 2007, 7, 244–251. [Google Scholar] [CrossRef] [PubMed]
- Stoodley, P.; Sauer, K.; Davies, D.G.; Costerton, J.W. Biofilms as complex differentiated communities. Annu. Rev. Microbiol. 2002, 56, 187–209. [Google Scholar] [CrossRef] [PubMed]
- O’Toole, G.; Kaplan, H.B.; Kolter, R. Biofilm formation as microbial development. Annu. Rev. Microbiol. 2000, 54, 49–79. [Google Scholar] [CrossRef] [PubMed]
- Sauer, K. The genomics and proteomics of biofilm formation. Genome Biol. 2003, 4, 219. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Purevdorj, B.; Costerton, J.W.; Stoodley, P. Influence of hydrodynamics and cell signaling on the structure and behavior of Pseudomonas aeruginosa biofilms. Appl. Environ. Microbiol. 2002, 68, 4457–4464. [Google Scholar] [CrossRef] [PubMed]
- Reimmann, C.; Ginet, N.; Michel, L.; Keel, C.; Michaux, P.; Krishnapillai, V.; Zala, M.; Heurlier, K.; Triandafillu, K.; Harms, H.; et al. Genetically programmed autoinducer destruction reduces virulence gene expression and swarming motility in Pseudomonas aeruginosa PAO1. Microbiology 2002, 148, 923–932. [Google Scholar] [CrossRef] [PubMed]
- Hentzer, M.; Eberl, L.; Givskov, M. Transcriptome analysis of Pseudomonas aeruginosa biofilm development: Anaerobic respiration and iron limitation. Biofilms 2005, 2, 37–61. [Google Scholar] [CrossRef]
- Flemming, H.; Wingender, J. The biofilm matrix. Nat. Rev. Microbiol. 2010, 8, 623–633. [Google Scholar] [CrossRef] [PubMed]
- Allesen-Holm, M.; Barken, K.B.; Yang, L.; Klausen, M.; Webb, J.S.; Kjelleberg, S.; Molin, S.; Givskov, M.; Tolker-Nielsen, T. A characterization of DNA release in Pseudomonas aeruginosa cultures and biofilms. Mol. Microbiol. 2006, 59, 1114–1128. [Google Scholar] [CrossRef] [PubMed]
- Whitchurch, C.B.; Tolker-Nielsen, T.; Ragas, P.C.; Mattick, J.S. Extracellular DNA Required for bacterial biofilm formation. Science 2002, 295, 1487. [Google Scholar] [CrossRef] [PubMed]
- Barken, K.B.; Pamp, S.J.; Yang, L.; Gjermansen, M.; Bertrand, J.J.; Klausen, M.; Givskov, M.; Whitchurch, C.B.; Engel, J.N.; Tolker-Nielsen, T. Roles of type IV pili, flagellum-mediated motility and extracellular DNA in the formation of mature multicellular structures in Pseudomonas aeruginosa biofilms. Environ. Microbiol. 2008, 10, 2331–2343. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Barken, K.B.; Skindersoe, M.E.; Christensen, A.B.; Givskov, M.; Tolker-Nielsen, T. Effects of iron on DNA release and biofilm development by Pseudomonas aeruginosa. Microbiology 2007, 153, 1318–1328. [Google Scholar] [CrossRef] [PubMed]
- Thomson, A.H. Human recombinant DNase in cystic fibrosis. J. R. Soc. Med. 1995, 88, 24–29. [Google Scholar] [PubMed]
- Ranasinha, C.; Assoufi, B.; Geddes, D.; Hodson, M.; Empey, D.; Shak, S.; Christiansen, D.; Fuchs, H. Efficacy and safety of short-term administration of aerosolised recombinant human DNase I in adults with stable stage cystic fibrosis. Lancet 1993, 342, 199–202. [Google Scholar] [CrossRef]
- Ramsey, B.W.; Astley, S.J.; Aitken, M.L.; Burke, W.; Colin, A.A.; Dorkin, H.L.; Eisenberg, J.D.; Gibson, R.L.; Harwood, I.R.; Schidlow, D.V.; et al. Efficacy and safety of short-term administration of aerosolized recombinant human deoxyribonuclease in patients with cystic fibrosis. Am. Rev. Respir. Dis. 1993, 148, 145–151. [Google Scholar] [CrossRef] [PubMed]
- Fuchs, H.J.; Borowitz, D.S.; Christiansen, D.H.; Morris, E.M.; Nash, M.L.; Ramsey, B.W.; Rosenstein, B.J.; Smith, A.L.; Wohl, M.E. Effect of Aerosolized recombinant human DNase on exacerbations of respiratory symptoms and on pulmonary function in patients with cystic fibrosis. N. Engl. J. Med. 1994, 331, 637–642. [Google Scholar] [CrossRef] [PubMed]
- Mathee, K.; Ciofu, O.; Sternberg, C.; Lindum, P.W.; Campbell, J.I.A.; Jensen, P.; Johnsen, A.H.; Givskov, M.; Ohman, D.E.; Molin, S.; et al. Mucoid conversion of Pseudomonas aeruginosa by hydrogen peroxide: A mechanism for virulence activation in the cystic fibrosis lung. Microbiology 1999, 145, 1349–1357. [Google Scholar] [CrossRef] [PubMed]
- Friedman, L.; Kolter, R. Two genetic loci produce distinct carbohydrate-rich structural components of the Pseudomonas aeruginosa biofilm matrix. J. Bacteriol. 2004, 186, 4457–4465. [Google Scholar] [CrossRef] [PubMed]
- Vasseur, P.; Vallet-Gely, I.; Soscia, C.; Genin, S.; Filloux, A. The pel genes of the Pseudomonas aeruginosa PAK strain are involved at early and late stages of biofilm formation. Microbiology 2005, 151, 985–997. [Google Scholar] [CrossRef] [PubMed]
- Colvin, K.M.; Gordon, V.D.; Murakami, K.; Borlee, B.R.; Wozniak, D.J.; Wong, G.C.L.; Parsek, M.R. The pel polysaccharide can serve a structural and protective role in the biofilm matrix of Pseudomonas aeruginosa. PLoS Pathog. 2011, 7, e1001264. [Google Scholar] [CrossRef] [PubMed]
- Coulon, C.; Vinogradov, E.; Filloux, A.; Sadovskaya, I. Chemical analysis of cellular and extracellular carbohydrates of a biofilm-forming strain Pseudomonas aeruginosa PA14. PLoS ONE 2010, 5, e14220. [Google Scholar] [CrossRef] [PubMed]
- Byrd, M.S.; Sadovskaya, I.; Vinogradov, E.; Lu, H.; Sprinkle, A.B.; Richardson, S.H.; Ma, L.; Ralston, B.; Parsek, M.R.; Anderson, E.M.; et al. Genetic and biochemical analyses of the Pseudomonas aeruginosa Psl exopolysaccharide reveal overlapping roles for polysaccharide synthesis enzymes in Psl and LPS production. Mol. Microbiol. 2009, 73, 622–638. [Google Scholar] [CrossRef] [PubMed]
- Colvin, K.M.; Irie, Y.; Tart, C.S.; Urbano, R.; Whitney, J.C.; Ryder, C.; Howell, P.L.; Wozniak, D.J.; Parsek, M.R. The Pel and Psl polysaccharides provide Pseudomonas aeruginosa structural redundancy within the biofilm matrix. Environ. Microbiol. 2012, 14, 1913–1928. [Google Scholar] [CrossRef] [PubMed]
- Schleheck, D.; Barraud, N.; Klebensberger, J.; Webb, J.S.; McDougald, D.; Rice, S.A.; Kjelleberg, S. Pseudomonas aeruginosa PAO1 preferentially grows as aggregates in liquid batch cultures and disperses upon starvation. PLoS ONE 2009, 4, e5513. [Google Scholar] [CrossRef] [PubMed]
- Sauer, K.; Cullen, M.C.; Rickard, A.H.; Zeef, L.A.H.; Davies, D.G.; Gilbert, P. Characterization of nutrient-induced dispersion in Pseudomonas aeruginosa PAO1 biofilm. J. Bacteriol. 2004, 186, 7312–7326. [Google Scholar] [CrossRef] [PubMed]
- Barraud, N.; Hassett, D.J.; Hwang, S.H.; Rice, S.A.; Kjelleberg, S.; Webb, J.S. Involvement of nitric oxide in biofilm dispersal of Pseudomonas aeruginosa. J. Bacteriol. 2006, 188, 7344–7353. [Google Scholar] [CrossRef] [PubMed]
- Banin, E.; Brady, K.M.; Greenberg, E.P. Chelator-induced dispersal and killing of Pseudomonas aeruginosa cells in a biofilm. Appl. Environ. Microbiol. 2006, 72, 2064–2069. [Google Scholar] [CrossRef] [PubMed]
- Romling, U.; Galperin, M.Y.; Gomelsky, M. Cyclic di-GMP: The first 25 years of a universal bacterial second messenger. Microbiol. Mol. Biol. Rev. 2013, 77, 1–52. [Google Scholar] [CrossRef] [PubMed]
- Schuster, M.; Lostroh, C.P.; Ogi, T.; Greenberg, E.P. Identification, timing, and signal specificity of Pseudomonas aeruginosa quorum-controlled genes: A transcriptome analysis. J. Bacteriol. 2003, 185, 2066–2079. [Google Scholar] [CrossRef] [PubMed]
- Wagner, V.E.; Bushnell, D.; Passador, L.; Brooks, A.I.; Iglewski, B.H. Microarray analysis of Pseudomonas aeruginosa quorum-sensing regulons: Effects of growth phase and environment. J. Bacteriol. 2003, 185, 2080–2095. [Google Scholar] [CrossRef] [PubMed]
- Howe, T.R.; Iglewski, B.H. Isolation and characterization of alkaline protease-deficient mutants of Pseudomonas aeruginosa in vitro and in a mouse eye model. Infect. Immun. 1984, 43, 1058–1063. [Google Scholar] [PubMed]
- Woods, D.E.; Cryz, S.J.; Friedman, R.L.; Iglewski, B.H. Contribution of toxin A and elastase to virulence of Pseudomonas aeruginosa in chronic lung infections of rats. Infect. Immun. 1982, 36, 1223–1228. [Google Scholar] [PubMed]
- Vasil, M.L.; Grant, C.C.R.; Prince, R.W. Regulation of exotoxin A synthesis in Pseudomonas aeruginosa: Characterization of toxA-lacZ fusions in wild-type and mutant strains. Mol. Microbiol. 1989, 3, 371–381. [Google Scholar] [CrossRef] [PubMed]
- Nicas, T.I.; Frank, D.W.; Stenzel, P.; Lile, J.D.; Iglewski, B.H. Role of exoenzyme S in chronic Pseudomonas aeruginosa lung infections. Eur. J. Clin. Microbiol. 1985, 4, 175–179. [Google Scholar] [CrossRef] [PubMed]
- Cox, C.D. Effect of pyochelin on the virulence of Pseudomonas aeruginosa. Infect. Immun. 1982, 36, 17–23. [Google Scholar] [PubMed]
- Ochsner, U.A.; Reiser, J. Autoinducer-mediated regulation of rhamnolipid biosurfactant synthesis in Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. USA 1995, 92, 6424–6428. [Google Scholar] [CrossRef] [PubMed]
- Bjarnsholt, T.; Jensen, P.Ø.; Burmølle, M.; Hentzer, M.; Haagensen, J.A.J.; Hougen, H.P.; Calum, H.; Madsen, K.G.; Moser, C.; Molin, S.; et al. Pseudomonas aeruginosa tolerance to tobramycin, hydrogen peroxide and polymorphonuclear leukocytes is quorum-sensing dependent. Microbiology 2005, 151, 373–383. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Nilsson, M.; Gjermansen, M.; Givskov, M.; Tolker-Nielsen, T. Pyoverdine and PQS mediated subpopulation interactions involved in Pseudomonas aeruginosa biofilm formation. Mol. Microbiol. 2009, 74, 1380–1392. [Google Scholar] [CrossRef] [PubMed]
- Shak, S.; Capon, D.J.; Hellmiss, R.; Marsters, S.A.; Baker, C.L. Recombinant human DNase I reduces the viscosity of cystic fibrosis sputum. Proc. Natl. Acad. Sci. USA 1990, 87, 9188–9192. [Google Scholar] [CrossRef] [PubMed]
- Ramphal, R.; Lhermitte, M.; Filliat, M.; Roussel, P. The binding of anti-pseudomonal antibiotics to macromolecules from cystic fibrosis sputum. J. Antimicrob. Chemother. 1988, 22, 483–490. [Google Scholar] [CrossRef] [PubMed]
- Hunt, B.E.; Weber, A.; Berger, A.; Ramsey, B.; Smith, A.L. Macromolecular mechanisms of sputum inhibition of tobramycin activity. Antimicrob. Agents Chemother. 1995, 39, 34–39. [Google Scholar] [CrossRef] [PubMed]
- Drew, K.R.P.; Sanders, L.K.; Culumber, Z.W.; Zribi, O.; Wong, G.C.L. Cationic amphiphiles increase activity of aminoglycoside antibiotic tobramycin in the presence of airway polyelectrolytes. J. Am. Chem. Soc. 2009, 131, 486–493. [Google Scholar] [CrossRef] [PubMed]
- Mulcahy, H.; Charron-Mazenod, L.; Lewenza, S. Extracellular DNA chelates cations and induces antibiotic resistance in Pseudomonas aeruginosa biofilms. PLoS Pathog. 2008, 4, e1000213. [Google Scholar] [CrossRef] [PubMed]
- Walker, T.S.; Tomlin, K.L.; Worthen, G.S.; Poch, K.R.; Lieber, J.G.; Saavedra, M.T.; Fessler, M.B.; Malcolm, K.C.; Vasil, M.L.; Nick, J.A. Enhanced Pseudomonas aeruginosa biofilm development mediated by human neutrophils. Infect. Immun. 2005, 73, 3693–3701. [Google Scholar] [CrossRef] [PubMed]
- Chiang, W.C.; Nilsson, M.; Jensen, P.Ø.; Høiby, N.; Nielsen, T.E.; Givskov, M.; Tolker-Nielsen, T. Extracellular DNA shields against aminoglycosides in Pseudomonas aeruginosa biofilms. Antimicrob. Agents Chemother. 2013, 57, 2352–2361. [Google Scholar] [CrossRef] [PubMed]
- Mahajan-Miklos, S.; Tan, M.W.; Rahme, L.G.; Ausubel, F.M. Molecular mechanisms of bacterial virulence elucidated using a Pseudomonas aeruginosa-Caenorhabditis elegans pathogenesis model. Cell 1999, 96, 47–56. [Google Scholar] [CrossRef]
- Wilson, R.; Sykes, D.A.; Watson, D.; Rutman, A.; Taylor, G.W.; Cole, P.J. Measurement of Pseudomonas aeruginosa phenazine pigments in sputum and assessment of their contribution to sputum sol toxicity for respiratory epithelium. Infect. Immun. 1988, 56, 2515–2517. [Google Scholar] [PubMed]
- Nutman, J.; Chase, P.A.; Dearborn, D.G.; Berger, M.; Sorensen, R.U. Suppression of lymphocyte proliferation by Pseudomonas aeruginosa phenazine pigments. ISR J. Med. Sci. 1988, 24, 228–232. [Google Scholar] [PubMed]
- Britigan, B.E.; Railsback, M.A.; Cox, C.D. The Pseudomonas aeruginosa secretory product pyocyanin inactivates alpha1 protease inhibitor: Implications for the pathogenesis of cystic fibrosis lung disease. Infect. Immun. 1999, 67, 1207–1212. [Google Scholar] [PubMed]
- Denning, G.M.; Railsback, M.A.; Rasmussen, G.T.; Cox, C.D.; Britigan, B.E. Pseudomonas pyocyanine alters calcium signaling in human airway epithelial cells. Am. J. Physiol. 1998, 274, L893–L900. [Google Scholar] [PubMed]
- Kamath, J.M.; Britigan, B.E.; Cox, C.D.; Shasby, D.M. Pyocyanin from Pseudomonas aeruginosa inhibits prostacyclin release from endothelial cells. Infect. Immun. 1995, 63, 4921–4923. [Google Scholar] [PubMed]
- Mühlradt, P.F.; Tsai, H.; Conradt, P. Effects of pyocyanine, a blue pigment from Pseudomonas aeruginosa, on separate steps of T cell activation: Interleukin 2 (IL 2) production, IL 2 receptor formation, proliferation and induction of cytolytic activity. Eur. J. Immunol. 1986, 16, 434–440. [Google Scholar] [CrossRef] [PubMed]
- Das, T.; Manefield, M. Pyocyanin promotes extracellular DNA release in Pseudomonas aeruginosa. PLoS ONE 2012, 7, e46718. [Google Scholar] [CrossRef] [PubMed]
- Das, T.; Kutty, S.K.; Tavallaie, R.; Ibugo, A.I.; Panchompoo, J.; Sehar, S.; Aldous, L.; Yeung, A.W.S.; Thomas, S.R.; Kumar, N.; et al. Phenazine virulence factor binding to extracellular DNA is important for Pseudomonas aeruginosa biofilm. Sci. Rep. 2015, 5, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Maier, R.M.; Soberón-Chávez, G. Pseudomonas aeruginosa rhamnolipids: Biosynthesis and potential applications. Appl. Microbiol. Biotechnol. 2000, 54, 625–633. [Google Scholar] [CrossRef] [PubMed]
- Rendell, N.B.; Taylor, G.W.; Somerville, M.; Todd, H.; Wilson, R.; Cole, P.J. Characterisation of Pseudomonas rhamnolipids. Biochim. Biophys. Acta (BBA) Lipids Lipid Metab. 1990, 1045, 189–193. [Google Scholar] [CrossRef]
- Ochsner, U.A.; Koch, A.K.; Fiechter, A.; Reiser, J. Isolation and characterization of a regulatory gene affecting rhamnolipid biosurfactant synthesis in Pseudomonas aeruginosa. J. Bacteriol. 1994, 176, 2044–2054. [Google Scholar] [CrossRef] [PubMed]
- Rahim, R.; Ochsner, U.A.; Olvera, C.; Graninger, M.; Messner, P.; Lam, J.S.; Soberón-Chávez, G. Cloning and functional characterization of the Pseudomonas aeruginosa rhlC gene that encodes rhamnosyltransferase 2, an enzyme responsible for di-rhamnolipid biosynthesis. Mol. Microbiol. 2001, 40, 708–718. [Google Scholar] [CrossRef] [PubMed]
- Johnson, M.K.; Boese-Marrazzo, D. Production and properties of heat-stable extracellular hemolysin from Pseudomonas aeruginosa. Infect. Immun. 1980, 29, 1028–1033. [Google Scholar] [PubMed]
- Jensen, P.; Bjarnsholt, T.; Phipps, R.; Rasmussen, T.B.; Calum, H.; Christoffersen, L.; Moser, C.; Williams, P.; Pressler, T.; Givskov, M.; et al. Rapid necrotic killing of polymorphonuclear leukocytes is caused by quorum-sensing-controlled production of rhamnolipid by Pseudomonas aeruginosa. Microbiology 2007, 153, 1329–1338. [Google Scholar] [CrossRef] [PubMed]
- McClure, C.D.; Schiller, N.L. Effects of Pseudomonas aeruginosa rhamnolipids on human monocyte-derived macrophages. J. Leukoc. Biol. 1992, 51, 97–102. [Google Scholar] [PubMed]
- Shryock, T.R.; Silver, S.A.; Banschbach, M.W.; Kramer, J.C. Effect of Pseudomonas aeruginosa rhamnolipid on human neutrophil migration. Curr. Microbiol. 1984, 10, 323–328. [Google Scholar] [CrossRef]
- Déziel, E.; Lépine, F.; Milot, S.; Villemur, R. rhlA is required for the production of a novel biosurfactant promoting swarming motility in Pseudomonas aeruginosa: 3-(3-Hydroxyalkanoyloxy)alkanoic acids (HAAs), the precursors of rhamnolipids. Microbiology 2003, 149, 2005–2013. [Google Scholar] [CrossRef] [PubMed]
- Davey, M.E.; Caiazza, N.C.; O’Toole, G.A. Rhamnolipid surfactant production affects biofilm architecture in Pseudomonas aeruginosa PAO1. J. Bacteriol. 2003, 185, 1027–1036. [Google Scholar] [CrossRef] [PubMed]
- Haba, E.; Pinazo, A.; Jauregui, O.; Espuny, M.J.; Infante, M.R.; Manresa, A. Physicochemical characterization and antimicrobial properties of rhamnolipids produced by Pseudomonas aeruginosa 47T2 NCBIM 40044. Biotechnol. Bioeng. 2003, 81, 316–322. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Soberón-Chávez, G.; Aguirre-Ramírez, M.; Sánchez, R. The Pseudomonas aeruginosa RhlA enzyme is involved in rhamnolipid and polyhydroxyalkanoate production. J. Ind. Microbiol. Biotechnol. 2005, 32, 675. [Google Scholar] [CrossRef] [PubMed]
- Read, R.C.; Roberts, P.; Munro, N.; Rutman, A.; Hastie, A.; Shryock, T.; Hall, R.; McDonald-Gibson, W.; Lund, V.; Taylor, G. Effect of Pseudomonas aeruginosa rhamnolipids on mucociliary transport and ciliary beating. J. Appl. Physiol. 1992, 72, 2271–2277. [Google Scholar] [PubMed]
- Zulianello, L.; Canard, C.; Köhler, T.; Caille, D.; Lacroix, J.S.; Meda, P. Rhamnolipids are virulence factors that promote early infiltration of primary human airway epithelia by Pseudomonas aeruginosa. Infect. Immun. 2006, 74, 3134–3147. [Google Scholar] [CrossRef] [PubMed]
- Bjarnsholt, T.; Givskov, M. Quorum sensing inhibitory drugs as next generation antimicrobials: Worth the effort? Curr. Infect. Dis. Rep. 2008, 10, 22–28. [Google Scholar] [CrossRef] [PubMed]
- Alhede, M.; Bjarnsholt, T.; Jensen, P.; Phipps, R.K.; Moser, C.; Christophersen, L.; Christensen, L.D.; van Gennip, M.; Parsek, M.; Høiby, N.; et al. Pseudomonas aeruginosa recognizes and responds aggressively to the presence of polymorphonuclear leukocytes. Microbiology 2009, 155, 3500–3508. [Google Scholar] [CrossRef] [PubMed]
- Morici, L.A.; Carterson, A.J.; Wagner, V.E.; Frisk, A.; Schurr, J.R.; Zu Bentrup, K.H.; Hassett, D.J.; Iglewski, B.H.; Sauer, K.; Schurr, M.J. Pseudomonas aeruginosa AlgR represses the Rhl quorum-sensing system in a biofilm-specific manner. J. Bacteriol. 2007, 189, 7752–7764. [Google Scholar] [CrossRef] [PubMed]
- Van Gennip, M.; Christensen, L.D.; Alhede, M.; Phipps, R.; Jensen, P.Ø.; Christophersen, L.; Pamp, S.J.; Moser, C.; Mikkelsen, P.J.; Koh, A.Y.; et al. Inactivation of the rhlA gene in Pseudomonas aeruginosa prevents rhamnolipid production, disabling the protection against polymorphonuclear leukocytes. APMIS 2009, 117, 537–546. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.; Wu, J.; Deng, Y.; Wang, J.; Wang, C.; Wang, J.; Chang, C.; Dong, Y.; Williams, P.; Zhang, L.-H. A cell-cell communication signal integrates quorum sensing and stress response. Nat. Chem. Biol. 2013, 9, 339–343. [Google Scholar] [CrossRef] [PubMed]
- Ciofu, O.; Mandsberg, L.F.; Bjarnsholt, T.; Wassermann, T.; Høiby, N. Genetic adaptation of Pseudomonas aeruginosa during chronic lung infection of patients with cystic fibrosis: Strong and weak mutators with heterogeneous genetic backgrounds emerge in mucA and/or lasR mutants. Microbiology 2010, 156, 1108–1119. [Google Scholar] [CrossRef] [PubMed]
- Hoffman, L.R.; Kulasekara, H.D.; Emerson, J.; Houston, L.S.; Burns, J.L.; Ramsey, B.W.; Miller, S.I. Pseudomonas aeruginosa lasR mutants are associated with cystic fibrosis lung disease progression. J. Cyst. Fibros. 2009, 8, 66–70. [Google Scholar] [CrossRef] [PubMed]
- Smith, E.E.; Buckley, D.G.; Wu, Z.; Saenphimmachak, C.; Hoffman, L.R.; D’Argenio, D.A.; Miller, S.I.; Ramsey, B.W.; Speert, D.P.; Moskowitz, S.M.; et al. V Genetic adaptation by Pseudomonas aeruginosa to the airways of cystic fibrosis patients. Proc. Natl. Acad. Sci. USA 2006, 103, 8487–8492. [Google Scholar] [CrossRef] [PubMed]
- D’Argenio, D.A.; Wu, M.; Hoffman, L.R.; Kulasekara, H.D.; Déziel, E.; Smith, E.E.; Nguyen, H.; Ernst, R.K.; Larson Freeman, T.J.; Spencer, D.H.; et al. Growth phenotypes of Pseudomonas aeruginosa lasR mutants adapted to the airways of cystic fibrosis patients. Mol. Microbiol. 2007, 64, 512–533. [Google Scholar] [CrossRef] [PubMed]
- Bjarnsholt, T.; Jensen, P.Ø.; Jakobsen, T.H.; Phipps, R.; Nielsen, A.K.; Rybtke, M.T.; Tolker-Nielsen, T.; Givskov, M.; Høiby, N.; Ciofu, O. Quorum sensing and virulence of Pseudomonas aeruginosa during lung infection of cystic fibrosis patients. PLoS ONE 2010, 5, e10115. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Römling, U.; Amikam, D. Cyclic di-GMP as a second messenger. Curr. Opin. Microbiol. 2006, 9, 218–228. [Google Scholar] [CrossRef] [PubMed]
- Galperin, M.Y. Bacterial signal transduction network in a genomic perspective. Environ. Microbiol. 2004, 6, 552–567. [Google Scholar] [CrossRef] [PubMed]
- Hengge, R. Principles of c-di-GMP signalling in bacteria. Nat. Rev. Microbiol. 2009, 7, 263–273. [Google Scholar] [CrossRef] [PubMed]
- Christen, M.; Christen, B.; Folcher, M.; Schauerte, A.; Jenal, U. Identification and characterization of a cyclic di-GMP-specific phosphodiesterase and its allosteric control by GTP. J. Biol. Chem. 2005, 280, 30829–30837. [Google Scholar] [CrossRef] [PubMed]
- Ryan, R.P.; Fouhy, Y.; Lucey, J.F.; Crossman, L.C.; Spiro, S.; He, Y.-W.; Zhang, L.-H.; Heeb, S.; Cámara, M.; Williams, P.; et al. Cell-cell signaling in Xanthomonas campestris involves an HD-GYP domain protein that functions in cyclic di-GMP turnover. Proc. Natl. Acad. Sci. USA 2006, 103, 6712–6717. [Google Scholar] [CrossRef] [PubMed]
- Kulasakara, H.; Lee, V.; Brencic, A.; Liberati, N.; Urbach, J.; Miyata, S.; Lee, D.G.; Neely, A.N.; Hyodo, M.; Hayakawa, Y.; et al. Analysis of Pseudomonas aeruginosa diguanylate cyclases and phosphodiesterases reveals a role for bis-(3′-5′)-cyclic-GMP in virulence. Proc. Natl. Acad. Sci. USA 2006, 103, 2839–2844. [Google Scholar] [CrossRef] [PubMed]
- Düvel, J.; Bertinetti, D.; Möller, S.; Schwede, F.; Morr, M.; Wissing, J.; Radamm, L.; Zimmermann, B.; Genieser, H.G.; Jänsch, L.; et al. A chemical proteomics approach to identify c-di-GMP binding proteins in Pseudomonas aeruginosa. J. Microbiol. Methods 2012, 88, 229–236. [Google Scholar] [CrossRef] [PubMed]
- Hay, I.D.; Remminghorst, U.; Rehm, B.H.A. MucR, a novel membrane-associated regulator of alginate biosynthesis in Pseudomonas aeruginosa. Appl. Environ. Microbiol. 2009, 75, 1110–1120. [Google Scholar] [CrossRef] [PubMed]
- Hickman, J.W.; Harwood, C.S. Identification of FleQ from Pseudomonas aeruginosa as a c-di-GMP-responsive transcription factor. Mol. Microbiol. 2008, 69, 376–389. [Google Scholar] [CrossRef] [PubMed]
- Lee, V.T.; Matewish, J.M.; Kessler, J.L.; Hyodo, M.; Hayakawa, Y.; Lory, S. A cyclic-di-GMP receptor required for bacterial exopolysaccharide production. Mol. Microbiol. 2007, 65, 1474–1484. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Meissner, A.; Wild, V.; Simm, R.; Rohde, M.; Erck, C.; Bredenbruch, F.; Morr, M.; Römling, U.; Häussler, S. Pseudomonas aeruginosa cupA-encoded fimbriae expression is regulated by a GGDEF and EAL domain-dependent modulation of the intracellular level of cyclic diguanylate. Environ. Microbiol. 2007, 9, 2475–2485. [Google Scholar] [CrossRef] [PubMed]
- Borlee, B.R.; Goldman, A.D.; Murakami, K.; Samudrala, R.; Wozniak, D.J.; Parsek, M.R. Pseudomonas aeruginosa uses a cyclic-di-GMP-regulated adhesin to reinforce the biofilm extracellular matrix. Mol. Microbiol. 2010, 75, 827–842. [Google Scholar] [CrossRef] [PubMed]
- Christensen, L.D.; van Gennip, M.; Rybtke, M.T.; Wu, H.; Chiang, W.C.; Alhede, M.; Høiby, N.; Nielsen, T.E.; Givskov, M.; Tolker-Nielsena, T. Clearance of Pseudomonas aeruginosa foreign-body biofilm infections through reduction of the cyclic di-gmp level in the bacteria. Infect. Immun. 2013, 81, 2705–2713. [Google Scholar] [CrossRef] [PubMed]
- Huang, B.; Whitchurch, C.B.; Mattick, J.S. FimX, a multidomain protein connecting environmental signals to twitching motility in Pseudomonas aeruginosa. J. Bacteriol. 2003, 185, 7068–7076. [Google Scholar] [CrossRef] [PubMed]
- Guzzo, C.R.; Salinas, R.K.; Andrade, M.O.; Farah, C.S. PILZ Protein Structure and Interactions with PILB and the FIMX EAL Domain: Implications for Control of Type IV Pilus Biogenesis. J. Mol. Biol. 2009, 393, 848–866. [Google Scholar] [CrossRef] [PubMed]
- Kulasekara, H.D.; Ventre, I.; Kulasekara, B.R.; Lazdunski, A.; Filloux, A.; Lory, S. A novel two-component system controls the expression of Pseudomonas aeruginosa fimbrial cup genes. Mol. Microbiol. 2005, 55, 368–380. [Google Scholar] [CrossRef] [PubMed]
- Mikkelsen, H.; Ball, G.; Giraud, C.; Filloux, A. Expression of Pseudomonas aeruginosa CupD Fimbrial genes is antagonistically controlled by RcsB and the EAL-containing PvrR response regulators. PLoS ONE 2009, 4, e6018. [Google Scholar] [CrossRef] [PubMed]
- Rao, F.; Yang, Y.; Qi, Y.; Liang, Z.-X. Catalytic mechanism of cyclic di-GMP-specific phosphodiesterase: A study of the EAL domain-containing RocR from Pseudomonas aeruginosa. J. Bacteriol. 2008, 190, 3622–3631. [Google Scholar] [CrossRef] [PubMed]
- D’Argenio, D.A.; Calfee, M.W.; Rainey, P.B.; Pesci, E.C. Autolysis and autoaggregation in Pseudomonas aeruginosa colony morphology mutants. J. Bacteriol. 2002, 184, 6481–6489. [Google Scholar] [CrossRef] [PubMed]
- Klebensberger, J.; Lautenschlager, K.; Bressler, D.; Wingender, J.; Philipp, B. Detergent-induced cell aggregation in subpopulations of Pseudomonas aeruginosa as a preadaptive survival strategy. Environ. Microbiol. 2007, 9, 2247–2259. [Google Scholar] [CrossRef] [PubMed]
- Klebensberger, J.; Birkenmaier, A.; Geffers, R.; Kjelleberg, S.; Philipp, B. SiaA and SiaD are essential for inducing autoaggregation as a specific response to detergent stress in Pseudomonas aeruginosa. Environ. Microbiol. 2009, 11, 3073–3086. [Google Scholar] [CrossRef] [PubMed]
- Irie, Y.; Starkey, M.; Edwards, A.N.; Wozniak, D.J.; Romeo, T.; Parsek, M.R. Pseudomonas aeruginosa biofilm matrix polysaccharide Psl is regulated transcriptionally by RpoS and post-transcriptionally by RsmA. Mol. Microbiol. 2010, 78, 158–172. [Google Scholar] [CrossRef] [PubMed]
- Hickman, J.W.; Tifrea, D.F.; Harwood, C.S. A chemosensory system that regulates biofilm formation through modulation of cyclic diguanylate levels. Proc. Natl. Acad. Sci. USA 2005, 102, 14422–14427. [Google Scholar] [CrossRef] [PubMed]
- Irie, Y.; Borlee, B.R.; O’Connor, J.R.; Hill, P.J.; Harwood, C.S.; Wozniak, D.J.; Parsek, M.R. Self-produced exopolysaccharide is a signal that stimulates biofilm formation in Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. USA 2012, 109, 20632–20636. [Google Scholar] [CrossRef] [PubMed]
- Remminghorst, U.; Rehm, B.H.A. Alg44, a unique protein required for alginate biosynthesis in Pseudomonas aeruginosa. FEBS Lett. 2006, 580, 3883–3888. [Google Scholar] [CrossRef] [PubMed]
- Amikam, D.; Galperin, M.Y. PilZ domain is part of the bacterial c-di-GMP binding protein. Bioinformatics 2006, 22, 3–6. [Google Scholar] [CrossRef] [PubMed]
- Merighi, M.; Lee, V.T.; Hyodo, M.; Hayakawa, Y.; Lory, S. The second messenger bis-(3′-5′)-cyclic-GMP and its PilZ domain-containing receptor Alg44 are required for alginate biosynthesis in Pseudomonas aeruginosa. Mol. Microbiol. 2007, 65, 876–895. [Google Scholar] [CrossRef] [PubMed]
- Oglesby, L.L.; Jain, S.; Ohman, D.E. Membrane topology and roles of Pseudomonas aeruginosa Alg8 and Alg44 in alginate polymerization. Microbiology 2008, 154, 1605–1615. [Google Scholar] [CrossRef] [PubMed]
- Starkey, M.; Hickman, J.H.; Ma, L.; Zhang, N.; de Long, S.; Hinz, A.; Palacios, S.; Manoil, C.; Kirisits, M.J.; Starner, T.D.; et al. Pseudomonas aeruginosa Rugose small-colony variants have adaptations that likely promote persistence in the cystic fibrosis lung. J. Bacteriol. 2009, 191, 3492–3503. [Google Scholar] [CrossRef] [PubMed]
- Malone, J.G.; Jaeger, T.; Spangler, C.; Ritz, D.; Spang, A.; Arrieumerlou, C.; Kaever, V.; Landmann, R.; Jenal, U. YfiBNR mediates cyclic di-GMP dependent small colony variant formation and persistence in Pseudomonas aeruginosa. PLoS Pathog. 2010, 6, e1000804. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hrabak, E.M.; Willis, D.K. The lemA gene required for pathogenicity of Pseudomonas syringae pv. syringae on bean is a member of a family of two-component regulators. J. Bacteriol. 1992, 174, 3011–3020. [Google Scholar] [CrossRef] [PubMed]
- Kay, E.; Humair, B.; Dénervaud, V.; Riedel, K.; Spahr, S.; Eberl, L.; Valverde, C.; Haas, D. Two GacA-dependent small RNAs modulate the quorum-sensing response in Pseudomonas aeruginosa. J. Bacteriol. 2006, 188, 6026–6033. [Google Scholar] [CrossRef] [PubMed]
- Lapouge, K.; Schubert, M.; Allain, F.H.T.; Haas, D. Gac/Rsm signal transduction pathway of γ-proteobacteria: From RNA recognition to regulation of social behaviour. Mol. Microbiol. 2008, 67, 241–253. [Google Scholar] [CrossRef] [PubMed]
- Pessi, G.; Williams, F.; Hindle, Z.; Heurlier, K.; Holden, M.T.G.; Cámara, M.; Haas, D.; Williams, P. The global posttranscriptional regulator RsmA modulates production of virulence determinants and N-acylhomoserine lactones in Pseudomonas aeruginosa. J. Bacteriol. 2001, 183, 6676–6683. [Google Scholar] [CrossRef] [PubMed]
- Reimmann, C.; Beyeler, M.; Latifi, A.; Winteler, H.; Foglino, M.; Lazdunski, A.; Haas, D. The global activator GacA of Pseudomonas aeruginosa PAO positively controls the production of the autoinducer N-butyryl-homoserine lactone and the formation of the virulence factors pyocyanin, cyanide, and lipase. Mol. Microbiol. 1997, 24, 309–319. [Google Scholar] [CrossRef] [PubMed]
- Marden, J.N.; Diaz, M.R.; Walton, W.G.; Gode, C.J.; Betts, L.; Urbanowski, M.L.; Redinbo, M.R.; Yahr, T.L.; Wolfgang, M.C. An unusual CsrA family member operates in series with RsmA to amplify posttranscriptional responses in Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. USA 2013, 110, 15055–15060. [Google Scholar] [CrossRef] [PubMed]
- Morris, E.R.; Hall, G.; Li, C.; Heeb, S.; Kulkarni, R.V.; Lovelock, L.; Silistre, H.; Messina, M.; Cámara, M.; Emsley, J.; et al. Structural rearrangement in an RsmA/CsrA Ortholog of Pseudomonas aeruginosa creates a dimeric RNA-binding protein, RsmN. Structure 2013, 21, 1659–1671. [Google Scholar] [CrossRef] [PubMed]
- Ventre, I.; Goodman, A.L.; Vallet-Gely, I.; Vasseur, P.; Soscia, C.; Molin, S.; Bleves, S.; Lazdunski, A.; Lory, S.; Filloux, A. Multiple sensors control reciprocal expression of Pseudomonas aeruginosa regulatory RNA and virulence genes. Proc. Natl. Acad. Sci. USA 2006, 103, 171–176. [Google Scholar] [CrossRef] [PubMed]
- Goodman, A.L.; Merighi, M.; Hyodo, M.; Ventre, I.; Filloux, A.; Lory, S. Direct interaction between sensor kinase proteins mediates acute and chronic disease phenotypes in a bacterial pathogen. Genes Dev. 2009, 23, 249–259. [Google Scholar] [CrossRef] [PubMed]
- Chambonnier, G.; Roux, L.; Redelberger, D.; Fadel, F.; Filloux, A.; Sivaneson, M.; de Bentzmann, S.; Bordi, C. The hybrid histidine kinase LadS forms a multicomponent signal transduction system with the GacS/GacA two-component system in Pseudomonas aeruginosa. PLoS Genet. 2016, 12, e1006032. [Google Scholar] [CrossRef] [PubMed]
- Broder, U.N.; Jaeger, T.; Jenal, U. LadS is a calcium-responsive kinase that induces acute-to-chronic virulence switch in Pseudomonas aeruginosa. Nat. Microbiol. 2016, 2, 16184. [Google Scholar] [CrossRef] [PubMed]
- Miller, C.L.; Romero, M.; Rajasekhar Karna, S.L.; Chen, T.; Heeb, S.; Leung, K.P. RsmW, Pseudomonas aeruginosa small non-coding RsmA-binding RNA upregulated in biofilm versus planktonic growth conditions. BMC Microbiol. 2016, 16, 155. [Google Scholar] [CrossRef] [PubMed]
- Brencic, A.; Lory, S. Determination of the regulon and identification of novel mRNA targets of Pseudomonas aeruginosa RsmA. Mol. Microbiol. 2009, 72, 612–632. [Google Scholar] [CrossRef] [PubMed]
- Potvin, E.; Lehoux, D.E.; Kukavica-Ibrulj, I.; Richard, K.L.; Sanschagrin, F.; Lau, G.W.; Levesque, R.C. In vivo functional genomics of Pseudomonas aeruginosa for high-throughput screening of new virulence factors and antibacterial targets. Environ. Microbiol. 2003, 5, 1294–1308. [Google Scholar] [CrossRef] [PubMed]
- Mougous, J.D.; Cuff, M.E.; Raunser, S.; Shen, A.; Zhou, M.; Gifford, C.A.; Goodman, A.L.; Joachimiak, G.; Ordonez, C.L.; Lory, S.; et al. A Virulence lous of Pseudomonas aeruginosa encodes a protein secretion apparatus. Science 2006, 312, 1526–1530. [Google Scholar] [CrossRef] [PubMed]
- Coggan, K.A.; Wolfgang, M.C. Global regulatory pathways and cross-talk control Pseudomonas aeruginosa environmental lifestyle and virulence phenotype. Curr. Issues Mol. Biol. 2012, 14, 47–70. [Google Scholar] [PubMed]
- Mikkelsen, H.; McMullan, R.; Filloux, A. The Pseudomonas aeruginosa reference strain PA14 displays increased virulence due to a mutation in ladS. PLoS ONE 2011, 6, e29113. [Google Scholar] [CrossRef] [PubMed]
- Jimenez, P.N.; Koch, G.; Thompson, J.A.; Xavier, K.B.; Cool, R.H.; Quax, W.J. The multiple signaling systems regulating virulence in Pseudomonas aeruginosa. Microbiol. Mol. Biol. Rev. 2012, 76, 46–65. [Google Scholar] [CrossRef] [PubMed]
- Frangipani, E.; Visaggio, D.; Heeb, S.; Kaever, V.; Cámara, M.; Visca, P.; Imperi, F. The Gac/Rsm and cyclic-di-GMP signalling networks coordinately regulate iron uptake in Pseudomonas aeruginosa. Environ. Microbiol. 2014, 16, 676–688. [Google Scholar] [CrossRef] [PubMed]
- Moscoso, J.A.; Mikkelsen, H.; Heeb, S.; Williams, P.; Filloux, A. The Pseudomonas aeruginosa sensor RetS switches Type III and Type VI secretion via c-di-GMP signalling. Environ. Microbiol. 2011, 13, 3128–3138. [Google Scholar] [CrossRef] [PubMed]
- Moscoso, J.A.; Jaeger, T.; Valentini, M.; Hui, K.; Jenal, U.; Filloux, A. The diguanylate cyclase SadC is a central player in Gac/Rsm-mediated biofilm formation in Pseudomonas aeruginosa. J. Bacteriol. 2014, 196, 4081–4088. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Rybtke, M.T.; Jakobsen, T.H.; Hentzer, M.; Bjarnsholt, T.; Givskov, M.; Tolker-Nielsen, T. Computer-aided identification of recognized drugs as Pseudomonas aeruginosa quorum-sensing inhibitors. Antimicrob. Agents Chemother. 2009, 53, 2432–2443. [Google Scholar] [CrossRef] [PubMed]
- Tan, S.Y.-Y.; Liu, Y.; Chua, S.L.; Vejborg, R.M.; Jakobsen, T.H.; Chew, S.C.; Li, Y.; Nielsen, T.E.; Tolker-Nielsen, T.; Yang, L.; et al. Comparative systems biology analysis to study the mode of action of the isothiocyanate compound iberin on Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 2014, 58, 6648–6659. [Google Scholar] [CrossRef] [PubMed]
- Rybtke, M.T.; Borlee, B.R.; Murakami, K.; Irie, Y.; Hentzer, M.; Nielsen, T.E.; Givskov, M.; Parsek, M.R.; Tolker-Nielsen, T. Fluorescence-based reporter for gauging cyclic Di-GMP levels in Pseudomonas aeruginosa. Appl. Environ. Microbiol. 2012, 78, 5060–5069. [Google Scholar] [CrossRef] [PubMed]
- Rumbaugh, K.P.; Griswold, J.A.; Iglewski, B.H.; Hamood, A.N. Contribution of quorum sensing to the virulence of Pseudomonas aeruginosa in burn wound infections. Infect. Immun. 1999, 67, 5854–5862. [Google Scholar] [PubMed]
- Smith, R.S.; Harris, S.G.; Phipps, R.; Iglewski, B. The Pseudomonas aeruginosa quorum-sensing molecule N-(3-oxododecanoyl)homoserine lactone contributes to virulence and induces inflammation in vivo. J. Bacteriol. 2002, 184, 1132–1139. [Google Scholar] [CrossRef] [PubMed]
- Pearson, J.P.; Feldman, M.; Iglewski, B.H.; Prince, A. Pseudomonas aeruginosa cell-to-cell signaling is required for virulence in a model of acute pulmonary infection. Infect. Immun. 2000, 68, 4331–4334. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.; Song, Z.; Givskov, M.; Doring, G.; Worlitzsch, D.; Mathee, K.; Rygaard, J.; Høiby, N. Pseudomonas aeruginosa mutations in lasI and rhlI quorum sensing systems result in milder chronic lung infection. Microbiology 2016, 2318, 34–1105. [Google Scholar] [CrossRef] [PubMed]
- Christensen, L.D.; Moser, C.; Jensen, P.; Rasmussen, T.B.; Christophersen, L.; Kjelleberg, S.; Kumar, N.; Høiby, N.; Givskov, M.; Bjarnsholt, T. Impact of Pseudomonas aeruginosa quorum sensing of biofilm persistence in an in vivo intraperitoneal foreign-body infection model. Microbiology 2007, 153, 2312–2320. [Google Scholar] [CrossRef] [PubMed]
- Xiao, G.; Déziel, E.; He, J.; Lépine, F.; Lesic, B.; Castonguay, M.H.; Milot, S.; Tampakaki, A.P.; Stachel, S.E.; Rahme, L.G. MvfR, a key Pseudomonas aeruginosa pathogenicity LTTR-class regulatory protein, has dual ligands. Mol. Microbiol. 2006, 62, 1689–1699. [Google Scholar] [CrossRef] [PubMed]
- Givskov, M.; de Nys, R.; Manefield, M.; Gram, L.; Maximilien, R.; Eberl, L.; Molin, S.Ø.; Steinberg, P.D.; Kjelleberg, S. Eukaryotic interference with homoserine lactone-mediated prokaryotic signalling. J. Bacteriol. 1996, 178, 6618–6622. [Google Scholar] [CrossRef] [PubMed]
- Rasmussen, T.B.; Skindersoe, M.E.; Bjarnsholt, T.; Phipps, R.K.; Christensen, K.B.; Jensen, P.O.; Andersen, J.B.; Koch, B.; Larsen, T.O.; Hentzer, M.; et al. Identity and effects of quorum-sensing inhibitors produced by Penicillium species. Microbiology 2005, 151, 1325–1340. [Google Scholar] [CrossRef] [PubMed]
- LaLonde, R.T.; Bu, L.; Henwood, A.; Fiumano, J.; Zhang, L. Bromine-, chlorine-, and mixed halogen-substituted 4-methyl-2(5H)-furanones: Synthesis and mutagenic effects of halogen and hydroxyl group replacements. Chem. Res. Toxicol. 1997, 10, 1427–1436. [Google Scholar] [CrossRef] [PubMed]
- Janecki, T.; Błaszczyk, E.; Studzian, K.; Różalski, M.; Krajewska, U.; Janecka, A. New stereocontrolled synthesis and biological evaluation of 5-(1′-hydroxyalkyl)-3-methylidenetetrahydro-2-furanones as Potential Cytotoxic Agents. J. Med. Chem. 2002, 45, 1142–1145. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.; Nam, N.-H.; You, Y.-J.; Ahn, B.-Z. Synthesis and cytotoxicity of 3,4-diaryl-2(5H)-furanones. Bioorg. Med. Chem. Lett. 2002, 12, 719–722. [Google Scholar] [CrossRef]
- Read, R.; Kumar, N. Production of Furanones. U.S. Patent 20070032666 A1, 8 February 2007. [Google Scholar]
- Galloway, W.R.J.D.; Hodgkinson, J.T.; Bowden, S.; Welch, M.; Spring, D.R. Applications of small molecule activators and inhibitors of quorum sensing in Gram-negative bacteria. Trends Microbiol. 2017, 20, 449–458. [Google Scholar] [CrossRef] [PubMed]
- Passador, L.; Tucker, K.D.; Guertin, K.R.; Journet, M.P.; Kende, A.S.; Iglewski, B.H. Functional analysis of the Pseudomonas aeruginosa autoinducer PAI. J. Bacteriol. 1996, 178, 5995–6000. [Google Scholar] [CrossRef] [PubMed]
- Ishida, T.; Ikeda, T.; Takiguchi, N.; Kuroda, A.; Ohtake, H.; Kato, J. Inhibition of quorum sensing in Pseudomonas aeruginosa by N-acyl cyclopentylamides. Appl. Environ. Microbiol. 2007, 73, 3183–3188. [Google Scholar] [CrossRef] [PubMed]
- Smith, K.M.; Bu, Y.; Suga, H. Induction and inhibition of Pseudomonas aeruginosa quorum sensing by synthetic autoinducer analogs. Chem. Biol. 2003, 10, 81–89. [Google Scholar] [CrossRef]
- Smith, K.M.; Bu, Y.; Suga, H. Library screening for synthetic agonists and antagonists of a Pseudomonas aeruginosa autoinducer. Chem. Biol. 2003, 10, 563–571. [Google Scholar] [CrossRef]
- Mattmann, M.E.; Blackwell, H.E. Small molecules that modulate quorum sensing and control virulence in Pseudomonas aeruginosa. J. Org. Chem. 2010, 75, 6737–6746. [Google Scholar] [CrossRef] [PubMed]
- Bottomley, M.J.; Muraglia, E.; Bazzo, R.; Carfì, A. Molecular insights into quorum sensing in the human pathogen Pseudomonas aeruginosa from the structure of the virulence regulator LasR bound to Its autoinducer. J. Biol. Chem. 2007, 282, 13592–13600. [Google Scholar] [CrossRef] [PubMed]
- Zou, Y.; Nair, S.K. Molecular basis for the recognition of structurally distinct autoinducer Mimics by the Pseudomonas aeruginosa LasR quorum-sensing signaling receptor. Chem. Biol. 2009, 16, 961–970. [Google Scholar] [CrossRef] [PubMed]
- Müh, U.; Schuster, M.; Heim, R.; Singh, A.; Olson, E.R.; Greenberg, E.P. Novel Pseudomonas aeruginosa quorum-sensing inhibitors identified in an ultra-high-throughput screen. Antimicrob. Agents Chemother. 2006, 50, 3674–3679. [Google Scholar] [CrossRef] [PubMed]
- Rasmussen, T.B.; Bjarnsholt, T.; Skindersoe, M.E.; Hentzer, M.; Kristoffersen, P.; Köte, M.; Nielsen, J.; Eberl, L.; Givskov, M. Screening for quorum-sensing inhibitors (QSI) by use of a novel genetic system, the QSI selector. J. Bacteriol. 2005, 187, 1799–1814. [Google Scholar] [CrossRef] [PubMed]
- Bjarnsholt, T.; Jensen, P.Ø.; Rasmussen, T.B.; Christophersen, L.; Calum, H.; Hentzer, M.; Hougen, H.P.; Rygaard, J.; Moser, C.; Eberl, L.; et al. Garlic blocks quorum sensing and promotes rapid clearing of pulmonary Pseudomonas aeruginosa infections. Microbiology 2005, 151, 3873–3880. [Google Scholar] [CrossRef] [PubMed]
- Persson, T.; Hansen, T.H.; Rasmussen, T.B.; Skinderso, M.E.; Givskov, M.; Nielsen, J. Rational design and synthesis of new quorum-sensing inhibitors derived from acylated homoserine lactones and natural products from garlic. Org. Biomol. Chem. 2005, 3, 253–262. [Google Scholar] [CrossRef] [PubMed]
- Pratt, D.A. Garlic and other alliums. The lore and the science. By eric block. Angew. Chem. Int. Ed. 2010, 49, 7162. [Google Scholar] [CrossRef]
- Block, E.; Ahmad, S.; Jain, M.K.; Crecely, R.W.; Apitz-Castro, R.; Cruz, M.R. The chemistry of alkyl thiosulfate esters. 8. (E,Z)-Ajoene: A potent antithrombotic agent from garlic. J. Am. Chem. Soc. 1984, 106, 8295–8296. [Google Scholar] [CrossRef]
- Jakobsen, T.H.; Bragason, S.K.; Phipps, R.K.; Christensen, L.D.; van Gennip, M.; Alhede, M.; Skindersoe, M.; Larsen, T.O.; Høiby, N.; Bjarnsholt, T.; et al. Food as a source for quorum sensing inhibitors: Iberin from horseradish revealed as a quorum sensing inhibitor of Pseudomonas aeruginosa. Appl. Environ. Microbiol. 2012, 78, 2410–2421. [Google Scholar] [CrossRef] [PubMed]
- Ganin, H.; Rayo, J.; Amara, N.; Levy, N.; Krief, P.; Meijler, M.M. Sulforaphane and erucin, natural isothiocyanates from broccoli, inhibit bacterial quorum sensing. Med. Chem. Commun. 2013, 4, 175–179. [Google Scholar] [CrossRef]
- Amara, N.; Mashiach, R.; Amar, D.; Krief, P.; Spieser, S.A.H.; Bottomley, M.J.; Aharoni, A.; Meijler, M.M. Covalent inhibition of bacterial quorum sensing. J. Am. Chem. Soc. 2009, 131, 10610–10619. [Google Scholar] [CrossRef] [PubMed]
- Fong, J.; Yuan, M.; Jakobsen, T.H.; Mortensen, K.T.; Delos Santos, M.M.S.; Chua, S.L.; Yang, L.; Tan, C.H.; Nielsen, T.E.; Givskov, M. Disulfide bond-containing ajoene Analogues As Novel Quorum Sensing Inhibitors of Pseudomonas aeruginosa. J. Med. Chem. 2017, 60, 215–227. [Google Scholar] [CrossRef] [PubMed]
- Jakobsen, T.H.; van Gennip, M.; Phipps, R.K.; Shanmugham, M.S.; Christensen, L.D.; Alhede, M.; Skindersoe, M.E.; Rasmussen, T.B.; Friedrich, K.; Uthe, F.; et al. Ajoene, a sulfur-rich molecule from garlic, inhibits genes controlled by quorum sensing. Antimicrob. Agents Chemother. 2012, 56, 2314–2325. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Manefield, M.; Rasmussen, T.B.; Henzter, M.; Andersen, J.B.; Steinberg, P.; Kjelleberg, S.; Givskov, M. Halogenated furanones inhibit quorum sensing through accelerated LuxR turnover. Microbiology 2002, 148, 1119–1127. [Google Scholar] [CrossRef] [PubMed]
- Jakobsen, T.H.; Warming, A.N.; Vejborg, R.M.; Moscoso, J.A.; Stegger, M.; Lorenzen, F.; Rybtke, M.; Andersen, J.B.; Petersen, R.; Andersen, P.S.; et al. A broad range quorum sensing inhibitor working through sRNA inhibition. Sci. Rep. 2017, in press. [Google Scholar] [CrossRef] [PubMed]
- Pérez-Martínez, I.; Haas, D. Azithromycin inhibits expression of the GacA-dependent small RNAs RsmY and RsmZ in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 2011, 55, 3399–3405. [Google Scholar] [CrossRef] [PubMed]
- Skindersoe, M.E.; Alhede, M.; Phipps, R.; Yang, L.; Jensen, P.O.; Rasmussen, T.B.; Bjarnsholt, T.; Tolker-Nielsen, T.; Høiby, N.; Givskov, M. Effects of antibiotics on quorum sensing in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 2008, 52, 3648–3663. [Google Scholar] [CrossRef] [PubMed]
- Sambanthamoorthy, K.; Sloup, R.E.; Parashar, V.; Smith, J.M.; Kim, E.E.; Semmelhack, M.F.; Neiditch, M.B.; Waters, C.M. Identification of small molecules that antagonize diguanylate cyclase enzymes to inhibit biofilm formation. Antimicrob. Agents Chemother. 2012, 56, 5202–5211. [Google Scholar] [CrossRef] [PubMed]
- Sambanthamoorthy, K.; Luo, C.; Pattabiraman, N.; Feng, X.; Koestler, B.; Waters, C.M.; Palys, T.J. Identification of small molecules inhibiting diguanylate cyclases to control bacterial biofilm development. Biofouling 2014, 30, 17–28. [Google Scholar] [CrossRef] [PubMed]
- Lieberman, O.J.; Orr, M.W.; Wang, Y.; Lee, V.T. High-throughput screening using the differential radial capillary action of ligand assay identifies ebselen as an inhibitor of diguanylate cyclases. ACS Chem. Biol. 2014, 9, 183–192. [Google Scholar] [CrossRef] [PubMed]
- Groizeleau, J.; Rybtke, M.; Andersen, J.B.; Berthelsen, J.; Liu, Y.; Yang, L.; Nielsen, T.E.; Kaever, V.; Givskov, M.; Tolker-Nielsen, T. The anti-cancerous drug doxorubicin decreases the c-di-GMP content in Pseudomonas aeruginosa but promotes biofilm formation. Microbiology 2016, 162, 1797–1807. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Watt, S.; Wang, J.; Nakayama, S.; Sayre, D.A.; Lam, Y.F.; Lee, V.T.; Sintim, H.O. Potent suppression of c-di-GMP synthesis via I-site allosteric inhibition of diguanylate cyclases with 2′-F-c-di-GMP. Bioorg. Med. Chem. 2013, 21, 4396–4404. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Y.; Tsuji, G.; Opoku-Temeng, C.; Sintim, H.O. Inhibition of P. aeruginosa c-di-GMP phosphodiesterase RocR and swarming motility by a benzoisothiazolinone derivative. Chem. Sci. 2016, 7, 6238–6244. [Google Scholar] [CrossRef]
- Christensen, L.D.; van Gennip, M.; Jakobsen, T.H.; Alhede, M.; Hougen, H.P.; Høiby, N.; Bjarnsholt, T.; Givskov, M. Synergistic antibacterial efficacy of early combination treatment with tobramycin and quorum-sensing inhibitors against Pseudomonas aeruginosa in an intraperitoneal foreign-body infection mouse model. J. Antimicrob. Chemother. 2012, 67, 1198–1206. [Google Scholar] [CrossRef] [PubMed]
- Brackman, G.; Cos, P.; Maes, L.; Nelis, H.J.; Coenye, T. Quorum sensing inhibitors increase the susceptibility of bacterial biofilms to antibiotics in vitro and in vivo. Antimicrob. Agents Chemother. 2011, 55, 2655–2661. [Google Scholar] [CrossRef] [PubMed]
- Høiby, N. Recent advances in the treatment of Pseudomonas aeruginosa infections in cystic fibrosis. BMC Med. 2011, 9, 32. [Google Scholar] [CrossRef] [PubMed]



| Compounds | Structure | AHL a | Pqs a | rsmY/rsmZ b | In Vivo c | References |
|---|---|---|---|---|---|---|
| Natural Sources | ||||||
| (1) Ajoene |  | + | + d | + | + | [178,180] |
| (2) Iberin |  | + | + | + | ÷ | [146,174] |
| (3) Penicillic acid |  | + | NI | ÷ d | NI | [155] |
| (4) Patulin |  | + | NI | ÷ d | + | [155] |
| Synthetic sources | ||||||
| (6) PD12 |  | + | NI | NI | NI | [168] |
| (7) V-06-018 |  | + | NI | NI | NI | [168] |
| (8) Isothiocyanates |  | + | NI | NI | NI | [176] |
| (9) C-30 |  | + | ÷ | ÷ d | + | [12] |
| Commercial drugs | ||||||
| (10) Azithromycin |  | + | NI | + | + | [181,182] |
| (11) Nifuroxazide |  | + | + | NI | NI | [145] |
| (12) Salicylic acid |  | + | + | NI | NI | [145] |
| Compound | Structure | WspR IC50 (μM) | References |
|---|---|---|---|
| DGC inhibitors | |||
| Non-nucleotide | |||
| (13) DI-4 |  | 8.17 | [183] |
| (14) DI-10 |  | 12.2 | [183] |
| (15) LP3134 |  | 44.9 | [184] |
| (16) LP3145 |  | 70.93 | [184] |
| (17) LP4010 |  | 102.4 | [184] |
| (18) Ebselen |  | 13.6 | [185] |
| (19) Doxorubicin |  | ÷ a | [186] |
| Nucleotide | |||
| (20) 2′-F-c-di-GMP |  | NI | [187] |
| PDE inhibitors | |||
| Non-nucleotide | |||
| (21) Compound |  | NI | [188] |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jakobsen, T.H.; Tolker-Nielsen, T.; Givskov, M. Bacterial Biofilm Control by Perturbation of Bacterial Signaling Processes. Int. J. Mol. Sci. 2017, 18, 1970. https://doi.org/10.3390/ijms18091970
Jakobsen TH, Tolker-Nielsen T, Givskov M. Bacterial Biofilm Control by Perturbation of Bacterial Signaling Processes. International Journal of Molecular Sciences. 2017; 18(9):1970. https://doi.org/10.3390/ijms18091970
Chicago/Turabian StyleJakobsen, Tim Holm, Tim Tolker-Nielsen, and Michael Givskov. 2017. "Bacterial Biofilm Control by Perturbation of Bacterial Signaling Processes" International Journal of Molecular Sciences 18, no. 9: 1970. https://doi.org/10.3390/ijms18091970
APA StyleJakobsen, T. H., Tolker-Nielsen, T., & Givskov, M. (2017). Bacterial Biofilm Control by Perturbation of Bacterial Signaling Processes. International Journal of Molecular Sciences, 18(9), 1970. https://doi.org/10.3390/ijms18091970