The Effects of Kaempferol-Inhibited Autophagy on Osteoclast Formation
Abstract
:1. Introduction
2. Results
2.1. Inhibitory Effects of Kaempferol on RAW 264.7 Cells
2.2. Kaempferol Inhibits RANKL-Induced Osteoclast Differentiation and Resorption in RAW 264.7 Cells
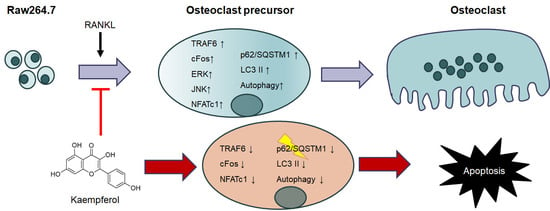
2.3. Inhibition of Osteoclastogenesis and Autophagy by Kaempferol
2.4. Kaempferol-Inhibited Autophagy Was Related to p62/SQSTM1 Degradation
3. Discussion
4. Materials and Methods
4.1. Reagents
4.2. Cell Culture
4.3. MTT Assay
4.4. Tartrate-Resistant Acid Phosphatase (TRAP) Staining
4.5. Osteoclast Pit Formation Assay
4.6. Western Blot Analysis
4.7. RNA Isolation and Real-Time PCR
4.8. Statistical Analysis
Acknowledgments
Author Contributions
Conflicts of Interest
Abbreviations
| BRONJ | Bisphosphonate related osteonecrosis of the jaws |
| BP | Bisphosphonates |
| TRAP | Tartrate-resistant acid phosphatase |
| MCSF | Macrophage colony-stimulating factor |
| RANK | Receptor activators of nuclear κB |
| RANKL | Receptor activators of nuclear κB ligand |
| TNF | Tumor necrosis factor |
| TRAF6 | TNF receptor-associated factor 6 |
| NFATc1 | Nuclear factor of activated T-cells, cytoplasmic 1 |
| CatK | Cathepsin K |
| DMSO | Dimethyl sulfoxide |
| PMSF | Phenylmethylsulfonyl fluoride |
| FBS | Fetal bovine serum |
| PVDF | Polyvinylidene difluoride membrane |
| MAPK | Mitogen-activated protein kinase |
| ERK | Extracellular signal-regulated kinase |
| JNK | c-Jun N-terminal kinase |
| PI-3K | Phosphatidylinositol 3-kinase |
| LC3 | Microtubule-associated protein 1A/1B-light chain 3 |
| ATG5 | Autophagy protein 5 |
| 3-MA | 3-Methyladenine |
| PARP | Poly ADP ribose polymerase |
References
- Boyle, W.J.; Simonet, W.S.; Lacey, D.L. Osteoclast differentiation and activation. Nature 2003, 423, 337–342. [Google Scholar] [CrossRef] [PubMed]
- Rachner, T.D.; Khosla, S.; Hofbauer, L.C. Osteoporosis: Now and the future. Lancet 2011, 377, 1276–1287. [Google Scholar] [CrossRef]
- Pang, J.L.; Ricupero, D.A.; Huang, S.; Fatma, N.; Singh, D.P.; Romero, J.R.; Chattopadhyay, N. Differential activity of kaempferol and quercetin in attenuating tumor necrosis factor receptor family signaling in bone cells. Biochem. Pharmacol. 2006, 71, 818–826. [Google Scholar] [CrossRef] [PubMed]
- Ikeda, K.; Takeshita, S. Factors and mechanisms involved in the coupling from bone resorption to formation: How osteoclasts talk to osteoblasts. J. Bone Metab. 2014, 21, 163–167. [Google Scholar] [CrossRef] [PubMed]
- Parfitt, A.M. Bone remodeling and bone loss: Understanding the pathophysiology of osteoporosis. Clin. Obstet. Gynecol. 1987, 30, 789–811. [Google Scholar] [CrossRef] [PubMed]
- Teitelbaum, S.L. Bone resorption by osteoclasts. Science 2000, 289, 1504–1508. [Google Scholar] [CrossRef] [PubMed]
- Pichardo, S.E.; Kuijpers, S.C.; van Merkesteyn, J.P. Bisphosphonate-related osteonecrosis of the jaws: Cohort study of surgical treatment results in seventy-four stage II/III patients. J. Craniomaxillofac. Surg. 2016, 44, 1216–1220. [Google Scholar] [CrossRef] [PubMed]
- Lazarovici, T.S.; Yahalom, R.; Taicher, S.; Schwartz-Arad, D.; Peleg, O.; Yarom, N. Bisphosphonate-Related Osteonecrosis of the Jaw Associated With Dental Implants. J. Oral Maxillofac. Surg. 2010, 68, 790–796. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.; Huang, S.; Guo, F.; Xu, F.; Cheng, P.; Ye, Y.; Dong, Y.; Xiang, W.; Chen, A. Dose-dependent inhibitory effects of zoledronic acid on osteoblast viability and function in vitro. Mol. Med. Rep. 2016, 13, 613–622. [Google Scholar] [CrossRef] [PubMed]
- Bloemen, V.; Schoenmaker, T.; De Vries, T.J.; Everts, V. Direct cell–cell contact between periodontal ligament fibroblasts and osteoclast precursors synergistically increases the expression of genes related to osteoclastogenesis. J. Cell. Physiol. 2010, 222, 565–573. [Google Scholar] [CrossRef] [PubMed]
- Jin, Z.; Wei, W.; Yang, M.; Du, Y.; Wan, Y. Mitochondrial complex I activity suppresses inflammation and enhances bone resorption by shifting macrophage-osteoclast polarization. Cell Metab. 2014, 20, 483–498. [Google Scholar] [CrossRef] [PubMed]
- Thomson, B.M.; Mundy, G.R.; Chambers, T.J. Tumor necrosis factors alpha and beta induce osteoblastic cells to stimulate osteoclastic bone resorption. J. Immunol. 1987, 138, 775–779. [Google Scholar] [PubMed]
- Kim, M.H.; Ryu, S.Y.; Bae, M.A.; Choi, J.-S.; Min, Y.K.; Kim, S.H. Baicalein inhibits osteoclast differentiation and induces mature osteoclast apoptosis. Food Chem. Toxicol. 2008, 46, 3375–3382. [Google Scholar] [CrossRef] [PubMed]
- Asai, K.; Funaba, M.; Murakami, M. Enhancement of RANKL-induced MITF-E expression and osteoclastogenesis by TGF-β. Cell Biochem. Funct. 2014, 32, 401–409. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Takayanagi, H. Osteoimmunology and the effects of the immune system on bone. Nat. Rev. Rheumatol. 2009, 5, 667–676. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Chen, G.; Zhang, W.; Xu, N.; Zhu, J.Y.; Jia, J.; Sun, Z.J.; Wang, Y.N.; Zhao, Y.F. Autophagy regulates hypoxia-induced osteoclastogenesis through the HIF-1α/BNIP3 signaling pathway. J. Cell. Physiol. 2012, 227, 639–648. [Google Scholar] [CrossRef] [PubMed]
- Chen, E.H.; Olson, E.N. Unveiling the mechanisms of cell-cell fusion. Science 2005, 308, 369–373. [Google Scholar] [CrossRef] [PubMed]
- Oberley, T.D.; Swanlund, J.M.; Zhang, H.J.; Kregel, K.C. Aging results in increased autophagy of mitochondria and protein nitration in rat hepatocytes following heat stress. J. Histochem. Cytochem. 2008, 56, 615–627. [Google Scholar] [CrossRef] [PubMed]
- Oikawa, T.; Kuroda, Y.; Matsuo, K. Regulation of osteoclasts by membrane-derived lipid mediators. Cell. Mol. Life Sci. 2013, 70, 3341–3353. [Google Scholar] [CrossRef] [PubMed]
- Ross, J.A.; Kasum, C.M. Dietary flavonoids: Bioavailability, metabolic effects, and safety. Ann. Rev. Nutr. 2002, 22, 19–34. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Pandey, A.K. Chemistry and Biological Activities of Flavonoids: An Overview. Sci. World J. 2013, 2013, 162750. [Google Scholar] [CrossRef] [PubMed]
- Rezk, B.M.; Haenen, G.R.M.M.; van der Vijgh, W.J.F.; Bast, A. The antioxidant activity of phloretin: The disclosure of a new antioxidant pharmacophore in flavonoids. Biochem. Biophys. Res. Commun. 2002, 295, 9–13. [Google Scholar] [CrossRef]
- Huang, L.; Yagura, T.; Chen, S. Sedative activity of hexane extract of Keampferia galanga L. and its active compounds. J. Ethnopharmacol. 2008, 120, 123–125. [Google Scholar] [CrossRef] [PubMed]
- Huang, W.-W.; Tsai, S.-C.; Peng, S.-F.; Lin, M.-W.; Chiang, J.-H.; Chiu, Y.-J.; Fushiya, S.; Tseng, M.T.; Yang, J.-S. Kaempferol induces autophagy through AMPK and AKT signaling molecules and causes G2/M arrest via downregulation of CDK1/cyclin B in SK-HEP-1 human hepatic cancer cells. Int. J. Oncol. 2013, 42, 2069–2077. [Google Scholar] [CrossRef] [PubMed]
- Wattel, A.; Kamel, S.; Mentaverri, R.; Lorget, F.; Prouillet, C.; Petit, J.-P.; Fardelonne, P.; Brazier, M. Potent inhibitory effect of naturally occurring flavonoids quercetin and kaempferol on in vitro osteoclastic bone resorption. Biochem. Pharmacol. 2003, 65, 35–42. [Google Scholar] [CrossRef]
- Miyake, M.; Arai, N.; Ushio, S.; Iwaki, K.; Ikeda, M.; Kurimoto, M. Promoting effect of kaempferol on the differentiation and mineralization of murine pre-osteoblastic cell line MC3T3-E1. Biosci. Biotechnol. Biochem. 2003, 67, 1199–1205. [Google Scholar] [CrossRef] [PubMed]
- Prouillet, C.; Maziere, J.C.; Maziere, C.; Wattel, A.; Brazier, M.; Kamel, S. Stimulatory effect of naturally occurring flavonols quercetin and kaempferol on alkaline phosphatase activity in MG-63 human osteoblasts through ERK and estrogen receptor pathway. Biochem. Pharmacol. 2004, 67, 1307–1313. [Google Scholar] [CrossRef] [PubMed]
- Guo, A.J.; Choi, R.C.; Zheng, K.Y.; Chen, V.P.; Dong, T.T.; Wang, Z.-T.; Vollmer, G.; Lau, D.T.; Tsim, K.W. Kaempferol as a flavonoid induces osteoblastic differentiation via estrogen receptor signaling. Chin. Med. 2012, 7, 10. [Google Scholar] [CrossRef] [PubMed]
- Xie, Z.; Klionsky, D.J. Autophagosome formation: Core machinery and adaptations. Nat. Cell Biol. 2007, 9, 1102–1109. [Google Scholar] [CrossRef] [PubMed]
- Ha, S.W.; Weitzmann, M.N.; Beck, G.R., Jr. Bioactive silica nanoparticles promote osteoblast differentiation through stimulation of autophagy and direct association with LC3 and p62. ACS Nano 2014, 8, 5898–5910. [Google Scholar] [CrossRef] [PubMed]
- Fleming, A.; Noda, T.; Yoshimori, T.; Rubinsztein, D.C. Chemical modulators of autophagy as biological probes and potential therapeutics. Nat. Chem. Biol. 2011, 7, 9–17. [Google Scholar] [CrossRef] [PubMed]
- Hippert, M.M.; O’Toole, P.S.; Thorburn, A. Autophagy in cancer: Good, bad, or both? Cancer Res. 2006, 66, 9349–9351. [Google Scholar] [CrossRef] [PubMed]
- Hocking, L.J.; Whitehouse, C.; Helfrich, M.H. Autophagy: A new player in skeletal maintenance? J. Bone Miner. Res. 2012, 27, 1439–1447. [Google Scholar] [CrossRef] [PubMed]
- Lee, W.S.; Lee, E.G.; Sung, M.S.; Yoo, W.H. Kaempferol inhibits IL-1beta-stimulated, RANKL-mediated osteoclastogenesis via downregulation of MAPKs, c-Fos, and NFATc1. Inflammation 2014, 37, 1221–1230. [Google Scholar] [CrossRef] [PubMed]
- Yamaguchi, M.; Hamamoto, R.; Uchiyama, S.; Ishiyama, K. Effects of flavonoid on calcium content in femoral tissue culture and parathyroid hormone-stimulated osteoclastogenesis in bone marrow culture in vitro. Mol. Cell. Biochem. 2007, 303, 83–88. [Google Scholar] [CrossRef] [PubMed]
- Cordoba, A.; Satue, M.; Gomez-Florit, M.; Hierro-Oliva, M.; Petzold, C.; Lyngstadaas, S.P.; Gonzalez-Martin, M.L.; Monjo, M.; Ramis, J.M. Flavonoid-Modified Surfaces: Multifunctional Bioactive Biomaterials with Osteopromotive, Anti-Inflammatory, and Anti-Fibrotic Potential. Adv. Healthc. Mater. 2015, 4, 540–549. [Google Scholar] [CrossRef] [PubMed]
- Middleton, E., Jr.; Kandaswami, C.; Theoharides, T.C. The effects of plant flavonoids on mammalian cells: Implications for inflammation, heart disease, and cancer. Pharmacol. Rev. 2000, 52, 673–751. [Google Scholar] [PubMed]
- Amic, D.; Davidovic-Amic, D.; Beslo, D.; Rastija, V.; Lucic, B.; Trinajstic, N. SAR and QSAR of the antioxidant activity of flavonoids. Curr. Med. Chem. 2007, 14, 827–845. [Google Scholar] [CrossRef] [PubMed]
- Galleano, M.; Calabro, V.; Prince, P.D.; Litterio, M.C.; Piotrkowski, B.; Vazquez-Prieto, M.A.; Miatello, R.M.; Oteiza, P.I.; Fraga, C.G. Flavonoids and metabolic syndrome. Ann. N. Y. Acad. Sci. 2012, 1259, 87–94. [Google Scholar] [CrossRef] [PubMed]
- Sharan, K.; Siddiqui, J.A.; Swarnkar, G.; Maurya, R.; Chattopadhyay, N. Role of phytochemicals in the prevention of menopausal bone loss: Evidence from in vitro and in vivo, human interventional and pharma-cokinetic studies. Curr. Med. Chem. 2009, 16, 1138–1157. [Google Scholar] [CrossRef] [PubMed]
- Bitto, A.; Polito, F.; Squadrito, F.; Marini, H.; D’Anna, R.; Irrera, N.; Minutoli, L.; Granese, R.; Altavilla, D. Genistein aglycone: A dual mode of action anti-osteoporotic soy isoflavone rebalancing bone turnover towards bone formation. Curr. Med. Chem. 2010, 17, 3007–3018. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Mediavilla, V.; Crespo, I.; Collado, P.S.; Esteller, A.; Sanchez-Campos, S.; Tunon, M.J.; Gonzalez-Gallego, J. The anti-inflammatory flavones quercetin and kaempferol cause inhibition of inducible nitric oxide synthase, cyclooxygenase-2 and reactive C-protein, and down-regulation of the nuclear factor kappaB pathway in Chang Liver cells. Eur. J. Pharmacol. 2007, 557, 221–229. [Google Scholar] [CrossRef] [PubMed]
- Noh, E.-M.; Kim, J.-S.; Hur, H.; Park, B.-H.; Song, E.-K.; Han, M.-K.; Kwon, K.-B.; Yoo, W.-H.; Shim, I.-K.; Lee, S. Cordycepin inhibits IL-1β-induced MMP-1 and MMP-3 expression in rheumatoid arthritis synovial fibroblasts. Rheumatology 2009, 48, 45–48. [Google Scholar] [CrossRef] [PubMed]
- Mizushima, N.; Komatsu, M. Autophagy: Renovation of cells and tissues. Cell 2011, 147, 728–741. [Google Scholar] [CrossRef] [PubMed]
- Levine, B.; Kroemer, G. Autophagy in the pathogenesis of disease. Cell 2008, 132, 27–42. [Google Scholar] [CrossRef] [PubMed]
- Liu, F.; Fang, F.; Yuan, H.; Yang, D.; Chen, Y.; Williams, L.; Goldstein, S.A.; Krebsbach, P.H.; Guan, J.-L. Suppression of autophagy by FIP200 deletion leads to osteopenia in mice through the inhibition of osteoblast terminal differentiation. J. Bone Miner. Res. 2013, 28, 2414–2430. [Google Scholar] [CrossRef] [PubMed]
- Hsu, H.; Lacey, D.L.; Dunstan, C.R.; Solovyev, I.; Colombero, A.; Timms, E.; Tan, H.-L.; Elliott, G.; Kelley, M.J.; Sarosi, I. Tumor necrosis factor receptor family member RANK mediates osteoclast differentiation and activation induced by osteoprotegerin ligand. Proc. Natl. Acad. Sci. USA 1999, 96, 3540–3545. [Google Scholar] [CrossRef] [PubMed]
- Gatica, D.; Chiong, M.; Lavandero, S.; Klionsky, D.J. Molecular Mechanisms of Autophagy in the Cardiovascular System. Circulation 2015, 116, 456–467. [Google Scholar] [CrossRef] [PubMed]
- Takayanagi, H.; Kim, S.; Koga, T.; Nishina, H.; Isshiki, M.; Yoshida, H.; Saiura, A.; Isobe, M.; Yokochi, T.; Inoue, J.-I. Induction and activation of the transcription factor NFATc1 (NFAT2) integrate RANKL signaling in terminal differentiation of osteoclasts. Dev. Cell 2002, 3, 889–901. [Google Scholar] [CrossRef]
- Ariyoshi, W.; Takahashi, T.; Kanno, T.; Ichimiya, H.; Takano, H.; Koseki, T.; Nishihara, T. Mechanisms involved in enhancement of osteoclast formation and function by low molecular weight hyaluronic acid. J. Biol. Chem. 2005, 280, 18967–18972. [Google Scholar] [CrossRef] [PubMed]
- Wattel, A.; Kamel, S.; Prouillet, C.; Petit, J.P.; Lorget, F.; Offord, E.; Brazier, M. Flavonoid quercetin decreases osteoclastic differentiation induced by RANKL via a mechanism involving NFκB and AP-1. J. Cell. Biochem. 2004, 92, 285–295. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.-Q.; Ovitt, C.; Grigoriadis, A.E.; Möhle-Steinlein, U.; Rüther, U.; Wagner, E.F. Bone and haematopoietic defects in mice lacking c-fos. Nature 1992, 360, 741–745. [Google Scholar] [CrossRef] [PubMed]
- Huang, H.; Chang, E.-J.; Ryu, J.; Lee, Z.H.; Lee, Y.; Kim, H.-H. Induction of c-Fos and NFATc1 during RANKL-stimulated osteoclast differentiation is mediated by the p38 signaling pathway. Biochem. Biophys. Res. Commun. 2006, 351, 99–105. [Google Scholar] [CrossRef] [PubMed]
- Arai, A.; Mizoguchi, T.; Harada, S.; Kobayashi, Y.; Nakamichi, Y.; Yasuda, H.; Penninger, J.M.; Yamada, K.; Udagawa, N.; Takahashi, N. c-Fos plays an essential role in the up-regulation of RANK expression in osteoclast precursors within the bone microenvironment. J. Cell Sci. 2012, 125, 2910–2917. [Google Scholar] [CrossRef] [PubMed]
- Ryter, S.W.; Mizumura, K.; Choi, A.M. The impact of autophagy on cell death modalities. Int. J. Cell Biol. 2014, 2014, 502676. [Google Scholar] [CrossRef] [PubMed]
- Maynard, A.A.; Dvorak, K.; Khailova, L.; Dobrenen, H.; Arganbright, K.M.; Halpern, M.D.; Kurundkar, A.R.; Maheshwari, A.; Dvorak, B. Epidermal growth factor reduces autophagy in intestinal epithelium and in the rat model of necrotizing enterocolitis. Am. J. Physiol. Gastrointest. Liver Physiol. 2010, 299, G614–G622. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.; Moon, J.Y.; Ahn, K.S.; Cho, S.K. Quercetin induces mitochondrial mediated apoptosis and protective autophagy in human glioblastoma U373MG cells. Oxid. Med. Cell. Longev. 2013, 2013, 596496. [Google Scholar] [CrossRef] [PubMed]





© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, C.-J.; Shin, S.-H.; Kim, B.-J.; Kim, C.-H.; Kim, J.-H.; Kang, H.-M.; Park, B.-S.; Kim, I.-R. The Effects of Kaempferol-Inhibited Autophagy on Osteoclast Formation. Int. J. Mol. Sci. 2018, 19, 125. https://doi.org/10.3390/ijms19010125
Kim C-J, Shin S-H, Kim B-J, Kim C-H, Kim J-H, Kang H-M, Park B-S, Kim I-R. The Effects of Kaempferol-Inhibited Autophagy on Osteoclast Formation. International Journal of Molecular Sciences. 2018; 19(1):125. https://doi.org/10.3390/ijms19010125
Chicago/Turabian StyleKim, Chang-Ju, Sang-Hun Shin, Bok-Joo Kim, Chul-Hoon Kim, Jung-Han Kim, Hae-Mi Kang, Bong-Soo Park, and In-Ryoung Kim. 2018. "The Effects of Kaempferol-Inhibited Autophagy on Osteoclast Formation" International Journal of Molecular Sciences 19, no. 1: 125. https://doi.org/10.3390/ijms19010125
APA StyleKim, C. -J., Shin, S. -H., Kim, B. -J., Kim, C. -H., Kim, J. -H., Kang, H. -M., Park, B. -S., & Kim, I. -R. (2018). The Effects of Kaempferol-Inhibited Autophagy on Osteoclast Formation. International Journal of Molecular Sciences, 19(1), 125. https://doi.org/10.3390/ijms19010125