Characterizing Metastatic HER2-Positive Gastric Cancer at the CDH1 Haplotype
Abstract
:1. Introduction
2. Results
2.1. Patient Characteristics
2.2. CDH1 Mutations
2.3. Association between CDH1 P7-Haplotype and mGC-HER2
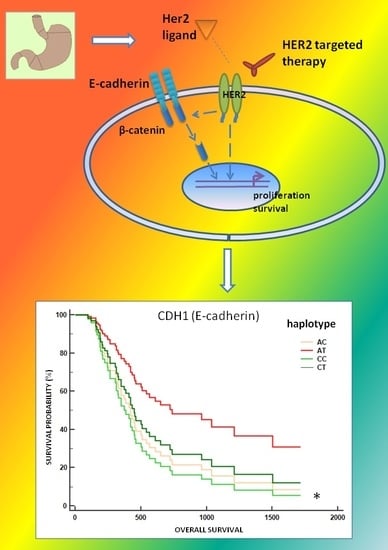
2.4. Association between the CDH1 P7 Haplotype and the Survival of mGC-HER2 Patients
3. Discussion
4. Materials and Methods
4.1. Study Population
4.2. Genotyping Analysis
4.3. Immunohistochemistry
4.4. Statistical Analysis
5. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
Abbreviations
| GC | Gastric cancer |
| mGC | Metastatic gastric cancer |
| HER2 | Human epidermal growth factor receptor 2 |
| E-cad | E-cadherin |
| IHC | Immunohistochemical |
| EBV | Epstein-barr virus |
| EMT | Epithelial-to-mesenchymal transition |
| MP | Metalloproteinase |
References
- Siegel, R.L.; Miller, K.D.; Jemal, A. Cancer statistics, 2017. CA Cancer J. Clin. 2017, 67, 7–30. [Google Scholar] [CrossRef] [PubMed]
- Santeufemia, D.A.; Lumachi, F.; Fadda, G.M.; Lo Re, G.; Miolo, G.; Basso, S.M.M.; Chiara, G.B.; Tumolo, S. Comment on “Repetitive transarterial chemoembolization (TACE) of liver metastases from gastric cancer: Local control and survival results”: Will there be clinical implications in the future? Eur. J. Radiol. 2013, 82, 1591–1592. [Google Scholar] [CrossRef] [PubMed]
- Romano, F.; Garancini, M.; Uggeri, F.; Degrate, L.; Nespoli, L.; Gianotti, L.; Nespoli, A.; Uggeri, F. Surgical treatment of liver metastases of gastric cancer: State of the art. World J. Surg. Oncol. 2012, 10, 157. [Google Scholar] [CrossRef] [PubMed]
- Catalano, V.; Graziano, F.; Santini, D.; D’Emidio, S.; Baldelli, A.M.; Rossi, D.; Vincenzi, B.; Giordani, P.; Alessandroni, P.; Testa, E.; et al. Second-line chemotherapy for patients with advanced gastric cancer: Who may benefit? Br. J. Cancer 2008, 99, 1402–1407. [Google Scholar] [CrossRef] [PubMed]
- Orditura, M.; Galizia, G.; Sforza, V.; Gambardella, V.; Fabozzi, A.; Laterza, M.M.; Andreozzi, F.; Ventriglia, J.; Savastano, B.; Mabilia, A.; et al. Treatment of gastric cancer. World J. Gastroenterol. 2014, 20, 1635–1649. [Google Scholar] [CrossRef] [PubMed]
- Bang, Y.-J.; Van Cutsem, E.; Feyereislova, A.; Chung, H.C.; Shen, L.; Sawaki, A.; Lordick, F.; Ohtsu, A.; Omuro, Y.; Satoh, T.; et al. Trastuzumab in combination with chemotherapy versus chemotherapy alone for treatment of HER2-positive advanced gastric or gastro-oesophageal junction cancer (ToGA): A phase 3, open-label, randomised controlled trial. Lancet 2010, 376, 687–697. [Google Scholar] [CrossRef]
- Garattini, S.K.; Basile, D.; Cattaneo, M.; Fanotto, V.; Ongaro, E.; Bonotto, M.; Negri, F.V.; Berenato, R.; Ermacora, P.; Cardellino, G.G.; et al. Molecular classifications of gastric cancers: Novel insights and possible future applications. World J. Gastrointest. Oncol. 2017, 9, 194–208. [Google Scholar] [CrossRef] [PubMed]
- Bonotto, M.; Garattini, S.K.; Basile, D.; Ongaro, E.; Fanotto, V.; Cattaneo, M.; Cortiula, F.; Iacono, D.; Cardellino, G.G.; Pella, N.; et al. Immunotherapy for gastric cancers: Emerging role and future perspectives. Expert Rev. Clin. Pharmacol. 2017, 10, 609–619. [Google Scholar] [CrossRef] [PubMed]
- Lubarsch, O.; Henke, F.; Rössle, R. Handbuch der Speziellen Pathologischen Anatomie und Histologie. Springer 1937, 9(part 3). Available online: http://www.springer.com/series/206 (accessed on 30 November 2017).
- Siewert, J.R.; Stein, H.J. Carcinoma of the gastroesophageal junction—Classification, pathology and extent of resection. Dis. Esophagus 1996, 9, 173–182. [Google Scholar] [CrossRef]
- WHO Classification of Tumours of the Digestive System, Fourth Edition. Available online: http://apps.who.int/bookorders/WHP/detart1.jsp?sesslan=1&codlan=1&codcol=70&codcch=4003 (accessed on 14 November 2017).
- Lauren, P. The two histological main types of gastric carcinoma: Diffuse and so-called intestinal-type carcinoma. An attempt at a histo-clinical classification. Acta Pathol. Microbiol. Scand. 1965, 64, 31–49. [Google Scholar] [CrossRef] [PubMed]
- Shan, L.; Ying, J.; Lu, N. HER2 expression and relevant clinicopathological features in gastric and gastroesophageal junction adenocarcinoma in a Chinese population. Diagn. Pathol. 2013, 8, 76. [Google Scholar] [CrossRef] [PubMed]
- The Cancer Genome Atlas Research Network. Comprehensive molecular characterization of gastric adenocarcinoma. Nature 2014, 513, 202–209. [Google Scholar] [CrossRef] [Green Version]
- Ye, X.S.; Yu, C.; Aggarwal, A.; Reinhard, C. Genomic alterations and molecular subtypes of gastric cancers in Asians. Chin. J. Cancer 2016, 35, 42. [Google Scholar] [CrossRef] [PubMed]
- Chu, C.-M.; Chen, C.-J.; Chan, D.-C.; Wu, H.-S.; Liu, Y.-C.; Shen, C.-Y.; Chang, T.-M.; Yu, J.; Harn, H.-J.; Yu, C.-P.; et al. CDH1 polymorphisms and haplotypes in sporadic diffuse and intestinal gastric cancer: A case–control study based on direct sequencing analysis. World J. Surg. Oncol. 2014, 12, 80. [Google Scholar] [CrossRef] [PubMed]
- Dang, H.-Z.; Yu, Y.; Jiao, S.-C. Prognosis of HER2 over-expressing gastric cancer patients with liver metastasis. World J. Gastroenterol. 2012, 18, 2402–2407. [Google Scholar] [CrossRef] [PubMed]
- Nami, B.; Wang, Z. HER2 in breast cancer stemness: A negative feedback loop towards trastuzumab resistance. Cancers 2017, 9, 40. [Google Scholar] [CrossRef] [PubMed]
- Namikawa, T.; Munekage, E.; Munekage, M.; Maeda, H.; Yatabe, T.; Kitagawa, H.; Sakamoto, K.; Obatake, M.; Kobayashi, M.; Hanazaki, K. Evaluation of a trastuzumab-containing treatment regimen for patients with unresectable advanced or recurrent gastric cancer. Mol. Clin. Oncol. 2016, 5, 74–78. [Google Scholar] [CrossRef] [PubMed]
- Caggiari, L.; Miolo, G.; Canzonieri, V.; De Zorzi, M.; Alessandrini, L.; Corona, G.; Cannizzaro, R.; Santeufemia, D.A.; Cossu, A.; Buonadonna, A.; et al. A new mutation of the CDH1 gene in a patient with an aggressive signet-ring cell carcinoma of the stomach. Cancer Biol. Ther. 2017, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Pisignano, G.; Napoli, S.; Magistri, M.; Mapelli, S.N.; Pastori, C.; Marco, S.D.; Civenni, G.; Albino, D.; Enriquez, C.; Allegrini, S.; et al. A promoter-proximal transcript targeted by genetic polymorphism controls E-cadherin silencing in human cancers. Nat. Commun. 2017, 8, 15622. [Google Scholar] [CrossRef] [PubMed]
- Zhan, Z.; Wu, J.; Zhang, J.-F.; Yang, Y.-P.; Tong, S.; Zhang, C.-B.; Li, J.; Yang, X.-W.; Dong, W. CDH1 gene polymorphisms, plasma CDH1 levels and risk of gastric cancer in a Chinese population. Mol. Biol. Rep. 2012, 39, 8107–8113. [Google Scholar] [CrossRef] [PubMed]
- Jiang, B.; Zhu, K.; Shao, H.; Bao, C.; Ou, J.; Sun, W. Lack of association between the CDH1 polymorphism and gastric cancer susceptibility: A meta-analysis. Sci. Rep. 2015, 5, 7891. [Google Scholar] [CrossRef] [PubMed]
- Chen, B.; Zhou, Y.; Yang, P.; Liu, L.; Qin, X.-P.; Wu, X.-T. CDH1 -160C>A gene polymorphism is an ethnicity-dependent risk factor for gastric cancer. Cytokine 2011, 55, 266–273. [Google Scholar] [CrossRef] [PubMed]
- Memni, H.; Macherki, Y.; Klayech, Z.; Ben-Haj-Ayed, A.; Farhat, K.; Remadi, Y.; Gabbouj, S.; Mahfoudh, W.; Bouzid, N.; Bouaouina, N.; et al. E-cadherin genetic variants predict survival outcome in breast cancer patients. J. Transl. Med. 2016, 14, 320. [Google Scholar] [CrossRef] [PubMed]
- Matsusaka, S.; Zhang, W.; Cao, S.; Hanna, D.L.; Sunakawa, Y.; Sebio, A.; Ueno, M.; Yang, D.; Ning, Y.; Parekh, A.; et al. TWIST1 polymorphisms predict survival in patients with metastatic colorectal cancer receiving first-line bevacizumab plus oxaliplatin-based chemotherapy. Mol. Cancer Ther. 2016, 15, 1405–1411. [Google Scholar] [CrossRef] [PubMed]
- Song, H.; Wang, X.; Yan, J.; Mi, N.; Jiao, X.; Hao, Y.; Zhang, W.; Gao, Y. Association of single-nucleotide polymorphisms of CDH1 with nonsyndromic cleft lip with or without cleft palate in a northern Chinese Han population. Medicine (Baltimore) 2017, 96, e5574. [Google Scholar] [CrossRef] [PubMed]
- Kluijt, I.; Siemerink, E.J.M.; Ausems, M.G.E.M.; van Os, T.A.M.; de Jong, D.; Simões-Correia, J.; van Krieken, J.H.; Ligtenberg, M.J.; Figueiredo, J.; van Riel, E.; et al. Dutch working group on hereditary gastric cancer CDH1-related hereditary diffuse gastric cancer syndrome: Clinical variations and implications for counseling. Int. J. Cancer 2012, 131, 367–376. [Google Scholar] [CrossRef] [PubMed]
- Benusiglio, P.R.; Malka, D.; Rouleau, E.; De Pauw, A.; Buecher, B.; Noguès, C.; Fourme, E.; Colas, C.; Coulet, F.; Warcoin, M.; et al. CDH1 germline mutations and the hereditary diffuse gastric and lobular breast cancer syndrome: A multicentre study. J. Med. Genet. 2013, 50, 486–489. [Google Scholar] [CrossRef] [PubMed]
- Frebourg, T.; Oliveira, C.; Hochain, P.; Karam, R.; Manouvrier, S.; Graziadio, C.; Vekemans, M.; Hartmann, A.; Baert-Desurmont, S.; Alexandre, C.; et al. Cleft lip/palate and CDH1/E-cadherin mutations in families with hereditary diffuse gastric cancer. J. Med. Genet. 2006, 43, 138–142. [Google Scholar] [CrossRef] [PubMed]
- Mossey, P.A.; Little, J.; Munger, R.G.; Dixon, M.J.; Shaw, W.C. Cleft lip and palate. Lancet 2009, 374, 1773–1785. [Google Scholar] [CrossRef]
- Pinheiro, H.; Bordeira-Carrico, R.; Seixas, S.; Carvalho, J.; Senz, J.; Oliveira, P.; Inacio, P.; Gusmao, L.; Rocha, J.; Huntsman, D.; et al. Allele-specific CDH1 downregulation and hereditary diffuse gastric cancer. Hum. Mol. Genet. 2010, 19, 943–952. [Google Scholar] [CrossRef] [PubMed]
- Repetto, O.; De Paoli, P.; De Re, V.; Canzonieri, V.; Cannizzaro, R. Levels of Soluble E-Cadherin in Breast, Gastric, and Colorectal Cancers. Available online: https://www.hindawi.com/journals/bmri/2014/408047/ (accessed on 17 November 2017).
- D’souza, B.; Taylor-Papadimitriou, J. Overexpression of ERBB2 in human mammary epithelial cells signals inhibition of transcription of the E-cadherin gene. Proc. Natl. Acad. Sci. USA 1994, 91, 7202–7206. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Chen, X.; Mao, Y.; Qu, Q.; Shen, K. Association of epithelial-mesenchymal transition with lapatinib resistance through multipe pathways activation in HER2-positive breast cancer. J. Clin. Oncol. 2014, 32, e11579. [Google Scholar] [CrossRef]
- Cheng, J.-C.; Qiu, X.; Chang, H.-M.; Leung, P.C.K. HER2 mediates epidermal growth factor-induced down-regulation of E-cadherin in human ovarian cancer cells. Biochem. Biophys. Res. Commun. 2013, 434, 81–86. [Google Scholar] [CrossRef] [PubMed]
- Garziera, M.; Canzonieri, V.; Cannizzaro, R.; Geremia, S.; Caggiari, L.; Zorzi, M.D.; Maiero, S.; Orzes, E.; Perin, T.; Zanussi, S.; et al. Identification and characterization of CDH1 germline variants in sporadic gastric cancer patients and in individuals at risk of gastric cancer. PLoS ONE 2013, 8, e77035. [Google Scholar] [CrossRef] [PubMed]
- Morcillo-Suarez, C.; Alegre, J.; Sangros, R.; Gazave, E.; de Cid, R.; Milne, R.; Amigo, J.; Ferrer-Admetlla, A.; Moreno-Estrada, A.; Gardner, M.; et al. SNP analysis to results (SNPator): A web-based environment oriented to statistical genomics analyses upon SNP data. Bioinformatics 2008, 24, 1643–1644. [Google Scholar] [CrossRef] [PubMed]
- Excoffier, L.; Laval, G.; Schneider, S. Arlequin (version 3.0): An integrated software package for population genetics data analysis. Evol. Bioinform. Online 2007, 1, 47–50. [Google Scholar] [CrossRef] [PubMed]



| CDH1 Region Gene | Reference Polymorphism | cDNA Change | Amino Acid Change | Type of Variant | Genotype | |||
|---|---|---|---|---|---|---|---|---|
| mGC-HER2 (n) | mGC (n) | |||||||
| Promoter | rs5030625 | c.-472delA | Polymorphic variant | G/A (1) | G/A (10) | |||
| Promoter | rs16260 | c.-285C>A | Polymorphic variant | A/A (4) | A/C (6) | A/A (3) | A/C (16) | |
| Promoter | rs34149581 | c.-276T>C | T/C (1) | |||||
| 5′UTR | rs34033771 | c.-71C>G | C/G (1) | |||||
| IV1 | rs3743674 | c.48+6C>T | Polymorphic variant | C/C (1) | T/C (9) | C/C (1) | ||
| EXON3 | rs1801023 | c.345G>A | p.Thr115= | Synonymous variant | G/A (1) | |||
| IV4 | rs33963999 | c.531+10G>C | G/C (2) | |||||
| IV5 | rs189969617 | c.688-14C>T | C/T (1) | |||||
| EXON7 | rs142822590 | c.892G>A | p.Ala298Thr | Missense variant | G/A (1) | |||
| EXON11 | SCV000588228.1 | c.1612delG | p.Asp538Thrfs*19 | Frameshift mutation | delG (1) | |||
| EXON12 | rs35187787 | c.1774G>A | p.Ala592Thr | Missense variant | G/A (1) | |||
| EXON12 | rs33969373 | c.1896C>T | p.HIS632= | Synonymous variant | C/T (2) | |||
| IV12 | rs2276330 | c.1937-13T>C | Polymorphic variant | C/T (2) | C/T (10) | |||
| EXON13 | rs1801552 | c.2076T>C | p.Ala692= | Polymorphic synonymous variant | C/T (5) | T/T (3) | C/T (24) | T/T (5) |
| IV13 | rs35686369 | c.2164+15_2164+16insA | insA (1) | insA (2) | ||||
| EXON14 | rs879026401 | c.2232A>G | p.Pro744= | Synonymous variant | A/G (1) | |||
| EXON14 | rs33964119 | c.2253C>T | p.Asn751= | Synonymous variant | C/T (1) | C/T (2) | ||
| EXON15 | rs587782549 | c.2204G>A | p.Arg796Gln | Missense variant | G/A (1) | |||
| Reference Polymorphism | Allele/Genotype | mGC-HER2 | Frequency | mGC | Frequency | p | OR (95% CI) |
|---|---|---|---|---|---|---|---|
| rs5030625 | |||||||
| Allele | G | 23 | 0.96 | 84 | 0.89 | 0.33 | 2.738 (0.33–22.51) |
| A | 1 | 0.04 | 10 | 0.11 | |||
| Genotype | G/G | 11 | 0.92 | 37 | 0.79 | ||
| G/A | 1 | 0.08 | 10 | 0.21 | |||
| A/A | 0 | 0.00 | 0 | 0.00 | |||
| Dominant model | GG/AA+AG | 11/1 | 0.92/0.08 | 37/10 | 0.79/0.21 | 0.30 | 2.973 (0.34–25.86) |
| Recessive model | AA/AG+GG | 0/12 | 0.00/1.00 | 0/47 | 0.00/1.00 | nv | |
| rs16260 | |||||||
| Allele | A | 14 | 0.58 | 22 | 0.23 | ≤0.001 | 4.582 (1.79–11.75) |
| C | 10 | 0.42 | 72 | 0.77 | |||
| Genotype | A/A | 4 | 0.33 | 3 | 0.06 | ||
| A/C | 6 | 0.50 | 16 | 0.34 | |||
| C/C | 2 | 0.17 | 28 | 0.60 | |||
| Recessive model | CC/AA+AC | 2/10 | 0.17/0.83 | 28/19 | 0.60/0.40 | ≤0.01 | 7.368 (1.45–37.46) |
| Dominant model | AA/AC+CC | 4/8 | 0.33/0.67 | 3/44 | 0.06/0.94 | 0.01 | 7.333 (1.37–39.18) |
| rs3743674 | |||||||
| Allele | T | 22 | 0.92 | 83 | 0.88 | 0.64 | 1.457 (0.30–7.07) |
| C | 2 | 0.08 | 11 | 0.12 | |||
| Genotype | T/T | 11 | 0.92 | 37 | 0.79 | ||
| T/C | 0 | 0.00 | 9 | 0.19 | |||
| C/C | 1 | 0.08 | 1 | 0.02 | |||
| Recessive model | CC/CT+TT | 1/11 | 0.08/0.92 | 1/46 | 0.02/0.98 | 0.29 | 4.182 (0.24–72.21) |
| Dominant model | TT/CC+CT | 11/1 | 0.92/0.08 | 37/10 | 0.79/0.21 | 0.30 | 2.973 (0.34–25.86) |
| rs2276330 | |||||||
| Allele | T | 22 | 0.92 | 84 | 0.90 | 0.74 | 1.309 (0.27–6.42) |
| C | 2 | 0.08 | 10 | 0.11 | |||
| Genotype | T/T | 10 | 0.83 | 37 | 0.79 | ||
| T/C | 2 | 0.17 | 10 | 0.21 | |||
| C/C | 0 | 0.00 | 0 | 0.00 | |||
| Dominant model | TT/CT+CC | 10/2 | 0.83/0.17 | 37/10 | 0.79/0.21 | 0.72 | 1.351 (0.25–7.19) |
| Recessive model | CC/TT+CT | 0/12 | 0.00/1.00 | 0/47 | 0.00/1.00 | nv | |
| rs1801552 | |||||||
| Allele | C | 13 | 0.54 | 60 | 0.64 | 0.39 | 0.670 (0.27–1.66) |
| T | 11 | 0.46 | 34 | 0.36 | |||
| C/C | 4 | 0.33 | 18 | 0.38 | |||
| Genotype | T/C | 5 | 0.42 | 24 | 0.51 | ||
| T/T | 3 | 0.25 | 5 | 0.11 | |||
| Recessive model | TT/CC+CT | 3/9 | 0.25/0.75 | 5/42 | 0.11/0.89 | 0.19 | 2.800 (0.56–13.90) |
| Dominant model | CC/CT+TT | 4/8 | 0.33/0.67 | 18/29 | 0.38/0.62 | 0.75 | 1.241 (0.33–4.72) |
| rs33964119 | |||||||
| C | 23 | 0.96 | 92 | 0.98 | 0.58 | 0.500 (0.04–5.76) | |
| Allele | T | 1 | 0.04 | 2 | 0.02 | ||
| C/C | 11 | 0.92 | 45 | 0.96 | |||
| Genotype | T/C | 1 | 0.08 | 2 | 0.04 | ||
| T/T | 0 | 0.00 | 0 | 0.00 | |||
| Recessive model | CC/CT+TT | 11/1 | 0.92/0.08 | 45/2 | 0.96/0.04 | 0.57 | 2.045 (0.17–24.66) |
| Dominant model | TT/CC+CT | 0/12 | 0.00/1.00 | 0/47 | 0.00/1.00 | nv | |
| Haplotype | mGC (N 94) | Frequency | mGC-HER2 (N 24) | Frequency | p | OR (95% CI) | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| rs5030625 | rs16260 | rs3743674 | rs2276330 | rs1801552 | rs33964119 | |||||||
| P1 | G | C | T | T | C | C | 19 | 0.20 | 2 | 0.08 | 0.24 | 0.359 (0.08–1.67) |
| P2 | G | A | T | T | C | C | 22 | 0.23 | 6 | 0.25 | 1.00 | 1.091 (0.39–3.09) |
| P3 | G | C | T | T | T | C | 31 | 0.33 | 4 | 0.17 | 0.14 | 0.406 (0.13–1.29) |
| P4 | G | C | T | C | C | C | 9 | 0.09 | 2 | 0.08 | 1.00 | 0.859 (0.17–4.26) |
| P5 | A | C | C | T | C | C | 7 | 0.07 | 1 | 0.04 | 0.69 | 0.540 (0.06–4.61) |
| P6 | A | C | C | C | C | C | 1 | 0.01 | 0 | 0.00 | 1.00 | |
| P7 | G | A | T | T | T | C | 0 | 0.00 | 7 | 0.29 | ≤0.001 | |
| P8 | G | C | C | T | T | C | 1 | 0.01 | 0 | 0.00 | 1.00 | |
| P9 | A | C | C | T | T | C | 2 | 0.02 | 0 | 0.00 | 1.00 | |
| P10 | G | C | T | T | C | T | 2 | 0.02 | 1 | 0.04 | 0.50 | 2.00 (0.17–23.03) |
| P11 | G | A | C | T | C | C | 0 | 0.00 | 1 | 0.04 | 0.20 | |
| Patient Identifier | Haplotype | CDH1 Germline Mutation | |||
|---|---|---|---|---|---|
| EXON11 c.1612delG | EXON12 c.1774G>A | IV13 c.2164+15_2164+16insA | EXON14 c.2253C>T | ||
| P287 | P5–P11 | ||||
| P291 | P2–P7 | ||||
| P292 | P4–P7 | ||||
| P296 | P2–P7 | ||||
| P297 | P3–P7 | ||||
| P301 | P2–P3 | ||||
| P303 | P2–P4 | ||||
| P380 | P1–P2 | ||||
| P391 | P2–P7 | ||||
| P486 | P7–P7 | ||||
| P582 | P3–P3 | ||||
| P586 | P1–P10 | ||||
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Caggiari, L.; Miolo, G.; Buonadonna, A.; Basile, D.; Santeufemia, D.A.; Cossu, A.; Palmieri, G.; De Zorzi, M.; Fornasarig, M.; Alessandrini, L.; et al. Characterizing Metastatic HER2-Positive Gastric Cancer at the CDH1 Haplotype. Int. J. Mol. Sci. 2018, 19, 47. https://doi.org/10.3390/ijms19010047
Caggiari L, Miolo G, Buonadonna A, Basile D, Santeufemia DA, Cossu A, Palmieri G, De Zorzi M, Fornasarig M, Alessandrini L, et al. Characterizing Metastatic HER2-Positive Gastric Cancer at the CDH1 Haplotype. International Journal of Molecular Sciences. 2018; 19(1):47. https://doi.org/10.3390/ijms19010047
Chicago/Turabian StyleCaggiari, Laura, Gianmaria Miolo, Angela Buonadonna, Debora Basile, Davide A. Santeufemia, Antonio Cossu, Giuseppe Palmieri, Mariangela De Zorzi, Mara Fornasarig, Lara Alessandrini, and et al. 2018. "Characterizing Metastatic HER2-Positive Gastric Cancer at the CDH1 Haplotype" International Journal of Molecular Sciences 19, no. 1: 47. https://doi.org/10.3390/ijms19010047
APA StyleCaggiari, L., Miolo, G., Buonadonna, A., Basile, D., Santeufemia, D. A., Cossu, A., Palmieri, G., De Zorzi, M., Fornasarig, M., Alessandrini, L., Canzonieri, V., Lo Re, G., Puglisi, F., Steffan, A., Cannizzaro, R., & De Re, V. (2018). Characterizing Metastatic HER2-Positive Gastric Cancer at the CDH1 Haplotype. International Journal of Molecular Sciences, 19(1), 47. https://doi.org/10.3390/ijms19010047