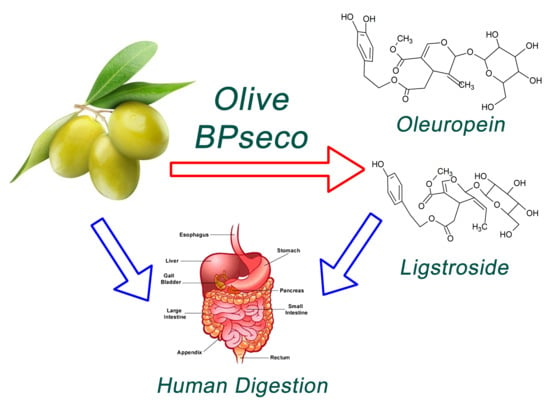
Probing Downstream Olive Biophenol Secoiridoids
Abstract
:1. Introduction
2. Results and Discussion
2.1. HPLC-MS Analysis
2.2. H3O+ and OH− Simulation and Characterization
2.3. Typed Neglected of Different Overlap (TNDO) Mapping
3. Materials and Methods
4. Conclusions
Author Contributions
Acknowledgments
Conflicts of Interest
Abbreviations
| AM1 | Austin model 1 |
| BPs | Biophenols |
| BPsecos | Biophenol secoiridoids |
| CAD | Collision activated dissociations |
| FW | Fresh weight |
| FMO | Frontier molecular orbital |
| Glu | Glucose |
| HOMO | highest occupied molecular orbital |
| HPLC-ESI-MS | High performance liquid chromatography electron spray ionization mass spectrometry |
| LUMO | Lowest unoccupied molecular orbital |
| Min | Minute |
| MS | Mass spectrometry |
| OLE | Oleuropein |
| PPO | Polyphenol oxidase |
| T | Time |
| TNDO | Typed neglected of different overlap |
References
- Sivakumar, G.; Uccella, N.A. Olive biophenols and conventional biotechnology from Mediterranean aliment culture. In Olives and Olive Oil in Health and Disease Prevention; Preedy, V.R., Watson, R.R., Eds.; Elsevier: New York, NY, USA, 2010; pp. 333–340. [Google Scholar]
- Crespo, M.C.; Tomé-Carneiro, J.; Dávalos, A.; Visioli, F. Pharma-nutritional properties of olive oil phenols. Transfer of new findings to Human nutrition. Foods 2018, 7, 90. [Google Scholar] [CrossRef] [PubMed]
- Sanchez-Rodriguez, E.; Lima-Cabello, E.; Biel-Glesson, S.; Fernandez-Navarro, J.R.; Calleja, M.; Roca, M.; Espejo-Calvo, J.A.; Gil-Extremera, B.; Soria-Florido, M.; de la Torre, R.; et al. Effects of virgin olive oils differing in their bioactive compound contents on metabolic syndrome and endothelial functional risk biomarkers in healthy adults: A randomized double-blind controlled trial. Nutrients 2018, 10, 626. [Google Scholar] [CrossRef] [PubMed]
- Nocella, C.; Cammisotto, V.; Fianchini, L.; D’Amico, A.; Novo, M.; Castellani, V.; Stefanini, L.; Violi, F.; Carnevale, R. Extra virgin olive oil and cardiovascular diseases: Benefits for Human health. Endocr. Metab. Immune Disord. Drug Targets 2018, 18, 4–13. [Google Scholar] [CrossRef] [PubMed]
- George, E.S.; Marshall, S.; Mayr, H.L.; Trakman, G.L.; Tatucu-Babet, O.A.; Lassemillante, A.M.; Bramley, A.; Reddy, A.J.; Forsyth, A.; Tierney, A.C.; Thomas, C.J.; Itsiopoulos, C.; Marx, W. The effect of high-polyphenol extra virgin olive oil on cardiovascular risk factors: A systematic review and meta-analysis. Crit. Rev. Food Sci. Nutr. 2018, 30, 1–138. [Google Scholar] [CrossRef] [PubMed]
- Imran, M.; Nadeem, M.; Gilani, S.A.; Khan, S.; Sajid, M.W.; Amir, R.M. Antitumor perspectives of oleuropein and its metabolite hydroxytyrosol: Recent updates. J. Food Sci. 2018, 83, 1781–1791. [Google Scholar] [CrossRef] [PubMed]
- Abtin, M.; Alivand, M.R.; Khaniani, M.S.; Bastami, M.; Zaeifizadeh, M.; Derakhshan, S.M. Simultaneous downregulation of miR-21 and miR-155 through oleuropein for breast cancer prevention and therapy. J. Cell. Biochem. 2018, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Kromhout, D.; Menotti, A.; Alberti-Fidanza, A.; Puddu, P.E.; Hollman, P.; Kafatos, A.; Tolonen, H.; Adachi, H.; Jacobs, D.R. Comparative ecologic relationships of saturated fat, sucrose, food groups, and a Mediterranean food pattern score to 50-year coronary heart disease mortality rates among 16 cohorts of the Seven Countries Study. Eur. J. Clin. Nutr. 2018, 72, 1103–1110. [Google Scholar] [CrossRef] [PubMed]
- Lockyer, S.; Rowland, I.; Spencer, J.P.E.; Yaqoob, P.; Stonehouse, W. Impact of phenolic-rich olive leaf extract on blood pressure, plasma lipids and inflammatory markers: A randomised controlled trial. Eur. J. Nutr. 2017, 56, 1421–1432. [Google Scholar] [CrossRef] [PubMed]
- Hanana, M.; Mezghenni, H.; Ben Ayed, R.; Ben Dhiab, A.; Jarradi, S.; Jamoussi, B.; Hamrouni, L. Nutraceutical potentialities of Tunisian Argan oil based on its physicochemical properties and fatty acid content as assessed through Bayesian network analyses. Lipids Health Dis. 2018, 17, 138. [Google Scholar] [CrossRef] [PubMed]
- Celano, R.; Piccinelli, A.L.; Pugliese, A.; Carabetta, S.; di Sanzo, R.; Rastrelli, L.; Russo, M. Insights into the analysis of phenolic secoiridoids in extra virgin olive oil. J. Agric. Food Chem. 2018, 66, 6053–6063. [Google Scholar] [CrossRef] [PubMed]
- Serra, G.; Incani, A.; Serreli, G.; Porru, L.; Melis, M.P.; Tuberoso, C.I.G.; Rossin, D.; Biasi, F.; Deiana, M. Olive oil polyphenols reduce oxysterols-induced redox imbalance and pro-inflammatory response in intestinal cells. Redox Biol. 2018, 17, 348–354. [Google Scholar] [CrossRef] [PubMed]
- Olivares-Vicente, M.; Barrajón-Catalán, E.; Herranz Lopez, M.; Segura Carretero, A.; Joven, J.; Encinar, J.A.; Micol, V. Plant-derived polyphenols in Human health: Biological activity, metabolites and putative molecular targets. Curr. Drug Metab. 2018, 19, 351–369. [Google Scholar] [CrossRef] [PubMed]
- Boss, A.; Bishop, K.S.; Marlow, G.; Barnett, M.P.; Ferguson, L.R. Evidence to support the anticancer effect of olive leaf extract and future directions. Nutrients 2016, 8, 513. [Google Scholar] [CrossRef] [PubMed]
- Sivakumar, G.; Briccoli Bati, C.; Uccella, N.A. HPLC-MS screening of the antioxidant profile of Italian olive cultivars. Chem. Nat. Compd. 2005, 41, 588–591. [Google Scholar] [CrossRef]
- Gentile, L.; Uccella, N.A.; Sivakumar, G. Soft-MS and computational mapping of oleuropein. Int. J. Mol. Sci. 2017, 18, 992. [Google Scholar] [CrossRef] [PubMed]
- Gentile, L.; Uccella, N.A.; Sivakumar, G. Oleuropein: Molecular dynamics and computation. Curr. Med. Chem. 2017, 24, 4315–4328. [Google Scholar] [CrossRef] [PubMed]
- Ambra, R.; Natella, F.; Bello, C.; Lucchetti, S.; Forte, V.; Pastore, G. Phenolics fate in table olives (Olea europaea L. cv. Nocellara del Belice) debittered using the Spanish and Castelvetrano methods. Food Res. Int. 2017, 100, 369–376. [Google Scholar] [CrossRef] [PubMed]
- Pino, A.; De Angelis, M.; Todaro, A.; Van Hoorde, K.; Randazzo, C.L.; Caggia, C. Fermentation of Nocellara Etnea table olives by functional starter cultures at different low salt concentrations. Front. Microbiol. 2018, 9, 1125. [Google Scholar] [CrossRef] [PubMed]
- Ji, S.T.; Kim, Y.J.; Jung, S.Y.; Kim, D.Y.; Kang, S.; Park, J.H.; Jang, W.B.; Ha, J.; Yun, J.; Kwon, S.M. Oleuropein attenuates hydrogen peroxide-induced autophagic cell death in human adipose-derived stem cells. Biochem. Biophys. Res. Commun. 2018, 499, 675–680. [Google Scholar] [CrossRef] [PubMed]
- Carnevale, R.; Silvestri, R.; Loffredo, L.; Novo, M.; Cammisotto, V.; Castellani, V.; Bartimoccia, S.; Nocella, C.; Violi, F. Oleuropein, a component of extra virgin olive oil, lowers postprandial glycaemia in healthy subjects. Br. J. Clin. Pharmacol. 2018, 84, 1566–1574. [Google Scholar] [CrossRef] [PubMed]
- Kerimi, A.; Nyambe-Silavwe, H.; Pyner, A.; Oladele, E.; Gauer, J.S.; Stevens, Y.; Williamson, G. Nutritional implications of olives and sugar: Attenuation of post-prandial glucose spikes in healthy volunteers by inhibition of sucrose hydrolysis and glucose transport by oleuropein. Eur. J. Nutr. 2018, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Sherif, I.O.; Al-Gayyar, M.M.H. Oleuropein potentiates anti-tumor activity of cisplatin against HepG2 through affecting proNGF/NGF balance. Life Sci. 2018, 198, 87–93. [Google Scholar] [CrossRef] [PubMed]
- Qabaha, K.; Al-Rimawi, F.; Qasem, A.; Naser, S.A. Oleuropein is responsible for the major Anti-inflammatory effects of olive leaf extract. J. Med. Food. 2018, 21, 302–305. [Google Scholar] [CrossRef] [PubMed]
- Cordero, J.G.; García-Escudero, R.; Avila, J.; Gargini, R.; García-Escudero, V. Benefit of oleuropein aglycone for Alzheimer’s disease by promoting autophagy. Oxid. Med. Cell. Longev. 2018, 2018, 5010741. [Google Scholar] [CrossRef] [PubMed]
- Palazzi, L.; Bruzzone, E.; Bisello, G.; Leri, M.; Stefani, M.; Bucciantini, M.; Polverino de Laureto, P. Oleuropein aglycone stabilizes the monomeric α-synuclein and favours the growth of non-toxic aggregates. Sci. Rep. 2018, 8, 8337. [Google Scholar] [CrossRef] [PubMed]
- Russo, N.; Capozzi, F.; Uccella, N.; Tocci, E.; Cremonini, M.A. Oleuropein biomimetic conformation by magnetic resonance experiments and molecular mechanics and dynamics. In Magnetic Resonance in Food Science. A View to the Future; Webb, G.A., Belton, P.S., Gil, A.M., Delgadillo, I., Eds.; Royal Society of Chemistry: Cambridge, UK, 2001; pp. 129–135. [Google Scholar]
- Demopoulos, V.; Karkoula, E.; Magiatis, P.; Melliou, E.; Kotsiras, A.; Mouroutoglou, C. Correlation of oleocanthal and oleacein concentration with pungency and bitterness in ‘Koroneiki’ virgin olive oil. Acta Hortic. 2015, 1099, 219–224. [Google Scholar] [CrossRef]
- Johnson, R.; Melliou, E.; Zweigenbaum, J.; Mitchell, A.E. Quantitation of oleuropein and related phenolics in cured Spanish-style green, California-style black ripe, and Greek-style natural fermentation olives. J. Agric. Food Chem. 2018, 66, 2121–2128. [Google Scholar] [CrossRef] [PubMed]
- Mougiou, N.; Trikka, F.; Trantas, E.; Ververidis, F.; Makris, A.; Argiriou, A.; Vlachonasios, K.E. Expression of hydroxytyrosol and oleuropein biosynthetic genes are correlated with metabolite accumulation during fruit development in olive, Olea europaea, cv. Koroneiki. Plant Physiol. Biochem. 2018, 128, 41–49. [Google Scholar] [CrossRef] [PubMed]
- Sivakumar, G.; Briccoli Bati, C.; Uccella, N. Demethyloleuropein and β-glucosidase activity in olive fruits. Biotechnol. J. 2007, 2, 381–385. [Google Scholar] [CrossRef] [PubMed]
- Velázquez-Palmero, D.; Romero-Segura, C.; García-Rodríguez, R.; Hernández, M.L.; Vaistij, F.E.; Graham, I.A.; Pérez, A.G.; Martínez-Rivas, J.M. An oleuropein β-glucosidase from olive fruit is involved in determining the phenolic composition of virgin olive oil. Front. Plant Sci. 2017, 8, 1902. [Google Scholar] [CrossRef] [PubMed]
- Afonso, C.; Cavaca, L.A.S.; Rodrigues, C.A.B.; Simeonov, S.P.; Gomes, R.F.A.; Coelho, J.A.S.; Romanelli, G.P.; Sathicq, A.G.; Martínez, J.J. Valorization of oleuropein via tunable acid-promoted methanolysis. ChemSusChem 2018, 11, 2300–2305. [Google Scholar] [CrossRef]
- Corominas-Faja, B.; Santangelo, E.; Cuyàs, E.; Micol, V.; Joven, J.; Ariza, X.; Segura-Carretero, A.; García, J.; Menendez, J.A. Computer-aided discovery of biological activity spectra for anti-aging and anti-cancer olive oil oleuropeins. Aging 2014, 6, 731–741. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kanakis, P.; Termentzi, A.; Michel, T.; Gikas, E.; Halabalaki, M.; Skaltsounis, AL. From olive drupes to olive oil. An HPLC-orbitrap-based qualitative and quantitative exploration of olive key metabolites. Planta Med. 2013, 79, 1576–1587. [Google Scholar] [CrossRef] [PubMed]
- Bianco, A.; Melchioni, C.; Ramunno, A.; Romeo, G.; Uccella, N.A. Phenolic components of Olea europaea—isolation of tyrosol derivatives. Nat. Prod. Res. 2004, 18, 29–32. [Google Scholar] [CrossRef] [PubMed]
- Borzillo, A.; Iannotta, N.; Uccella, N.A. Oinotria table olives: Quality evaluation during ripening and processing by biomolecular components. Eur. Food Res. Technol. 2000, 212, 113–121. [Google Scholar] [CrossRef]
- Enzo Perri, E.; Raffaelli, A.; Sindona, G. Quantitation of oleuropein in virgin olive oil by ionspray mass spectrometry−selected reaction monitoring. J. Agric Food Chem. 1999, 47, 4156–4160. [Google Scholar] [CrossRef]
- Bianco, A.; Uccella, N.A. Biophenolic components of olives. Food Res. Intern. 2000, 33, 475–485. [Google Scholar] [CrossRef]
- Del Coco, L.; Schena, F.P.; Fanizzi, F.P. 1H nuclear magnetic resonance study of olive oils commercially available as Italian products in the United States of America. Nutrients 2012, 4, 343–355. [Google Scholar] [CrossRef] [PubMed]
- Fabiani, R.; Sepporta, M.V.; Mazza, T.; Rosignoli, P.; Fuccelli, R.; De Bartolomeo, A.; Crescimanno, M.; Taticchi, A.; Esposto, S.; Servili, M.; et al. Influence of cultivar and concentration of selected phenolic constituents on the in vitro chemiopreventive potential of olive oil extracts. J. Agric. Food Chem. 2011, 59, 8167–8174. [Google Scholar] [CrossRef] [PubMed]
- Favati, F.; Condelli, N.; Galgano, F.; Caruso, M.C. Extra virgin olive oil bitterness evaluation by sensory and chemical analyses. Food Chem. 2013, 139, 949–954. [Google Scholar] [CrossRef] [PubMed]
- Rigacci, S.; Berti, A. Oleuropein aglycon: The bitter treasure of extra virgin olive oil. In Olive Oil and Health; Corrigan, J.D., Ed.; Nova eBook: New York, NY, USA, 2010; pp. 221–240. [Google Scholar]
- Mateos, R.; Cert, A.; Pérez-Camino, M.C.; García, J.M. Evaluation of virgin olive oil bitterness by quantification of secoiridoid derivatives. J. Am. Oil Chem. Soc. 2004, 81, 71–75. [Google Scholar] [CrossRef]
- Iaria, D.L.; Chiappetta, A.; Muzzalupo, I. A de novo transcriptomic approach to identify flavonoids and anthocyanins “switch-off” in olive (Olea europaea L.) drupes at different stages of maturation. Front. Plant Sci. 2016, 6, 1246. [Google Scholar] [CrossRef] [PubMed]
- Benincasa, C.; Muccilli, S.; Amenta, M.; Perri, E.; Romeo, F.V. Phenolic trend and hygienic quality of green table olives fermented with Lactobacillus plantarum starter culture. Food Chem. 2015, 186, 271–276. [Google Scholar] [CrossRef] [PubMed]
- Ciafardini, G.; Marsilio, V.; Lanza, B.; Pozzi, N. Hydrolysis of oleuropein by Lactobacillus plantarum strains associated with olive fermentation. Appl. Environ. Microbiol. 1994, 60, 4142–4147. [Google Scholar] [PubMed]
- Bianco, AD.; Jensen, S.R.; Olesen, J.; Passacantilli, P.; Ramunno, A. Acid rearrangement of secoiridoid related to oleuropein and secologanin. Eur. J. Org. Chem. 2003, 22, 4349–4354. [Google Scholar] [CrossRef]
- Procopio, A.; Alcaro, S.; Nardi, M.; Oliverio, M.; Ortuso, F.; Sacchetta, P.; Pieragostino, D.; Sindona, G. Synthesis, biological evaluation, and molecular modeling of oleuropein and its semi synthetic derivatives as cyclooxygenase inhibitors. J. Agric. Food Chem. 2009, 57, 11161–11167. [Google Scholar] [CrossRef] [PubMed]
- Brenes, M.; Garcia, A.; Garrido, A. Acid hydrolysis of secoiridoid aglycones during storage of virgin olive oil. J. Agric. Food Chem. 2001, 49, 5609–5614. [Google Scholar] [CrossRef] [PubMed]
- Capozzi, F.; Piperno, A.; Uccella, N.A. Oleuropein site selective hydrolysis by technomimetic nuclear magnetic resonance experiments. J. Agric. Food Chem. 2000, 48, 1623–1629. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.; Ma, F.; Ma, K.; Cao, L.; Zhao, L. Solvothermal synthesis and theoretical study of a polypyridium trimesylate adduct. J. Chem. Sci. 2011, 123, 687–696. [Google Scholar] [CrossRef]
- Deslongchamps, P.; Dory, Y.L.; Li, S. The relative rate of hydrolysis of a series of acyclic and six-membered cyclic acetals, ketals, othoesters, and orthocarbonates. Tetrahedron 2000, 56, 3533–3537. [Google Scholar] [CrossRef]
- Capasso, R.; Evidente, A.; Visca, C.; Gianfreda, L.; Maremonti, M.; Greco, G. Production of glucose and bioactive aglycone by chemical and enzymatic hydrolysis of purified oleuropein from Olea europea. Appl. Biochem. Biotech. 1996, 61, 365–377. [Google Scholar] [CrossRef]
- Edgecombe, S.C.; Stretch, G.L.; Hayball, P.J. Oleuropein, an antioxidant polyphenol from olive oil, is poorly absorbed from isolated perfused rat intestine. J. Nutr. 2000, 130, 2996–3002. [Google Scholar] [CrossRef] [PubMed]
- Jerman Klen, T.; Golc Wondra, A.; Vrhovšek, U.; Sivilotti, P.; Vodopivec, B.M. Olive fruit phenols transfer, transformation, and partition trail during laboratory-scale olive oil processing. J. Agric. Food Chem. 2015, 63, 4570–4579. [Google Scholar] [CrossRef] [PubMed]
- Polini, B.; Digiacomo, M.; Carpi, S.; Bertini, S.; Gado, F.; Saccomanni, G.; Macchia, M.; Nieri, P.; Manera, C.; Fogli, S. Oleocanthal and oleacein contribute to the in vitro therapeutic potential of extra virgin oil-derived extracts in non-melanoma skin cancer. Toxicol. In Vitro 2018, 52, 243–250. [Google Scholar] [CrossRef] [PubMed]
- Vissers, M.N.; Zock, P.L.; Roodenburg, A.J.; Leenen, R.; Katan, M.B. Olive oil phenols are absorbed in humans. J. Nutr. 2002, 132, 409–417. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez-Morató, J.; Robledo, P.; Tanner, J.A.; Boronat, A.; Pérez-Mañá, C.; Oliver Chen, C.Y.; Tyndale, R.F.; de la Torre, R. CYP2D6 and CYP2A6 biotransform dietary tyrosol into hydroxytyrosol. Food Chem. 2017, 217, 716–725. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Visioli, F.; Galli, C.; Bornet, F.; Mattei, A.; Patelli, R.; Galli, G.; Caruso, D. Olive oil phenolics are dose-dependently absorbed in humans. FEBS Lett. 2000, 468, 159–160. [Google Scholar] [CrossRef] [Green Version]
- Stupans, I.; Murray, M.; Kirlich, A.; Tuck, K.L.; Hayball, P.J. Inactivation of cytochrome P450 by the food-derived complex phenol oleuropein. Food Chem. Toxicol. 2001, 39, 1119–1124. [Google Scholar] [CrossRef]
- Young, D. Computational Chemistry: A Practical Guide for Applying Techniques to Real World Problems; John Wiley & Sons: New York, NY, USA, 2004. [Google Scholar]









| eV | OLE | OLE O2H+ | OLE GluO + H+ | OLEC11=O + H+ | H2O |
|---|---|---|---|---|---|
| HOMO | −10.94 | −12.69 | −12.58 | −12.83 | −17.82 |
| HOMO + 1 | −11.75 | −14.6 | −14.48 | −14.76 | −19.1 |
| HOMO + 2 | −12.13 | −15.21 | −15.04 | −14.98 | −20.68 |
| LUMO | 3.013 | −1.664 | −1.883 | −3.414 | 7.871 |
| LUMO-1 | 3.666 | −1.227 | −0.4203 | 0.3386 | 8.423 |
| LUMO-2 | 3.76 | −0.2421 | 0.1457 | 0.6718 | – |
| ∆E HOMO/UMO | 7.93 | 11.026 | 10.697 | 9.416 | – |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sivakumar, G.; Uccella, N.A.; Gentile, L. Probing Downstream Olive Biophenol Secoiridoids. Int. J. Mol. Sci. 2018, 19, 2892. https://doi.org/10.3390/ijms19102892
Sivakumar G, Uccella NA, Gentile L. Probing Downstream Olive Biophenol Secoiridoids. International Journal of Molecular Sciences. 2018; 19(10):2892. https://doi.org/10.3390/ijms19102892
Chicago/Turabian StyleSivakumar, Ganapathy, Nicola A. Uccella, and Luigi Gentile. 2018. "Probing Downstream Olive Biophenol Secoiridoids" International Journal of Molecular Sciences 19, no. 10: 2892. https://doi.org/10.3390/ijms19102892
APA StyleSivakumar, G., Uccella, N. A., & Gentile, L. (2018). Probing Downstream Olive Biophenol Secoiridoids. International Journal of Molecular Sciences, 19(10), 2892. https://doi.org/10.3390/ijms19102892