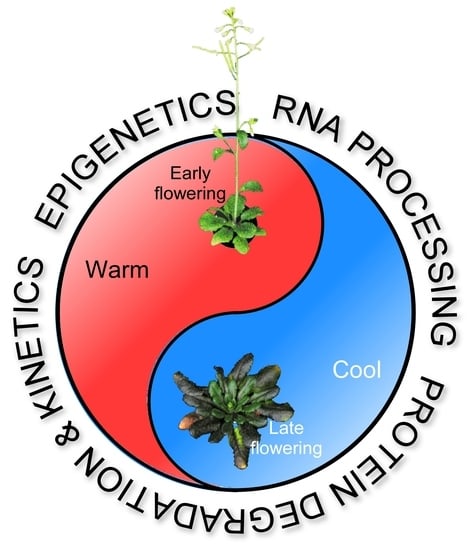
Ambient Temperature-Responsive Mechanisms Coordinate Regulation of Flowering Time
Abstract
:1. Introduction
2. Histone Variants, Histone Modification, and DNA Methylation
3. Non-Coding RNAs
3.1. Long Non-Coding RNAs (lncRNAs)
3.2. miRNAs
4. Alternative Splicing
5. 26S Proteasome-Dependent Protein Degradation
6. Thermal Reversion of Phytochromes
7. Circadian Clock Entrainment and Compensation as an Output of Temperature Perception
8. Plant Hormones as Secondary Signals in Temperature Perception
8.1. GAs, Key Flowering Time Regulators
8.2. BRs
8.3. Is JA Connected to Ambient Temperature-Mediated Flowering?
9. Conclusions and Future Perspectives
Author Contributions
Funding
Conflicts of Interest
References
- Hansen, J.; Sato, M.; Ruedy, R.; Lo, K.; Lea, D.W.; Medina-Elizade, M. Global temperature change. Proc. Natl. Acad. Sci. USA 2006, 103, 14288–14293. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cahill, N.; Rahmstorf, S.; Parnell, A.C. Change points of global temperature. Environ. Res. Lett. 2015, 10, 084002. [Google Scholar] [CrossRef] [Green Version]
- Pecl, G.T.; Araújo, M.B.; Bell, J.D.; Blanchard, J.; Bonebrake, T.C.; Chen, I.C.; Clark, T.D.; Colwell, R.K.; Danielsen, F.; Evengård, B.; et al. Biodiversity redistribution under climate change: Impacts on ecosystems and human well-being. Science 2017, 355, eaai9214. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pacifici, M.; Visconti, P.; Butchart, S.H.M.; Watson, J.E.M.; Cassola, F.M.; Rondinini, C. Species’ traits influenced their response to recent climate change. Nat. Clim. Chang. 2017, 7, 205–208. [Google Scholar] [CrossRef] [Green Version]
- Nicotra, A.B.; Atkin, O.K.; Bonser, S.P.; Davidson, A.M.; Finnegan, E.J.; Mathesius, U.; Poot, P.; Purugganan, M.D.; Richards, C.L.; Valladares, F.; et al. Plant phenotypic plasticity in a changing climate. Trends Plant Sci. 2010, 15, 684–692. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bouche, F.; Lobet, G.; Tocquin, P.; Perilleux, C. Flor-id: An interactive database of flowering-time gene networks in Arabidopsis thaliana. Nucleic Acids Res. 2016, 44, D1167–D1171. [Google Scholar] [CrossRef] [PubMed]
- Fitter, A.H. Rapid changes in flowering time in british plants. Science 2002, 296, 1689–1691. [Google Scholar] [CrossRef] [PubMed]
- Mohandass, D.; Zhao, J.-L.; Xia, Y.-M.; Campbell, M.J.; Li, Q.-J. Increasing temperature causes flowering onset time changes of alpine ginger roscoea in the central himalayas. J. Asia-Pac. Biodivers. 2015, 8, 191–198. [Google Scholar] [CrossRef]
- Liu, B.; Asseng, S.; Müller, C.; Ewert, F.; Elliott, J.; Lobell, D.B.; Martre, P.; Ruane, A.C.; Wallach, D.; Jones, J.W.; et al. Similar estimates of temperature impacts on global wheat yield by three independent methods. Nat. Clim. Chang. 2016, 6, 1130–1136. [Google Scholar] [CrossRef] [Green Version]
- Tacarindua, C.R.P.; Shiraiwa, T.; Homma, K.; Kumagai, E.; Sameshima, R. The effects of increased temperature on crop growth and yield of soybean grown in a temperature gradient chamber. Field Crop. Res. 2013, 154, 74–81. [Google Scholar] [CrossRef]
- Balasubramanian, S.; Sureshkumar, S.; Lempe, J.; Weigel, D. Potent induction of Arabidopsis thaliana flowering by elevated growth temperature. PLoS Genet. 2006, 2, e106. [Google Scholar] [CrossRef] [PubMed]
- Lempe, J.; Balasubramanian, S.; Sureshkumar, S.; Singh, A.; Schmid, M.; Weigel, D. Diversity of flowering responses in wild Arabidopsis thaliana strains. PLoS Genet. 2005, 1, e6. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Capovilla, G.; Schmid, M.; Pose, D. Control of flowering by ambient temperature. J. Exp. Bot. 2014, 66, 59–69. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Verhage, L.; Angenent, G.C.; Immink, R.G.H. Research on floral timing by ambient temperature comes into blossom. Trends Plant Sci. 2014, 19, 583–591. [Google Scholar] [CrossRef] [PubMed]
- Wigge, P.A. Ambient temperature signalling in plants. Curr. Opin. Plant Boil. 2013, 16, 661–666. [Google Scholar] [CrossRef] [PubMed]
- Penfield, S. Temperature perception and signal transduction in plants. New Phytol. 2008, 179, 615–628. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McClung, C.R.; Davis, S.J. Ambient thermometers in plants: From physiological outputs towards mechanisms of thermal sensing. Curr. Boil. 2010, 20, R1086–R1092. [Google Scholar] [CrossRef] [PubMed]
- Venkatesh, S.; Workman, J.L. Histone exchange, chromatin structure and the regulation of transcription. Nat. Rev. Mol. Cell Boil. 2015, 16, 178–189. [Google Scholar] [CrossRef] [PubMed]
- Luger, K.; Mäder, A.W.; Richmond, R.K.; Sargent, D.F.; Richmond, T.J. Crystal structure of the nucleosome core particle at 2.8 Å resolution. Nature 1997, 389, 251–260. [Google Scholar] [CrossRef] [PubMed]
- Kimura, H.; Cook, P.R. Kinetics of core histones in living human cells. J. Cell Boil. 2001, 153, 1341–1354. [Google Scholar] [CrossRef]
- Henikoff, S. Nucleosome destabilization in the epigenetic regulation of gene expression. Nat. Rev. Genet. 2008, 9, 15–26. [Google Scholar] [CrossRef] [PubMed]
- Jiang, D.; Berger, F. Histone variants in plant transcriptional regulation. Biochim. Biophys. Acta (BBA) Gene Regul. Mech. 2017, 1860, 123–130. [Google Scholar] [CrossRef] [PubMed]
- Henikoff, S.; Smith, M.M. Histone variants and epigenetics. Cold Spring Harb. Perspect. Boil. 2015, 7, a019364. [Google Scholar] [CrossRef] [PubMed]
- Talbert, P.B.; Henikoff, S. Environmental responses mediated by histone variants. Trends Cell Boil. 2014, 24, 642–650. [Google Scholar] [CrossRef] [PubMed]
- Jin, J.; Cai, Y.; Li, B.; Conaway, R.C.; Workman, J.L.; Conaway, J.W.; Kusch, T. In and out: Histone variant exchange in chromatin. Trends Biochem. Sci. 2005, 30, 680–687. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.V.; Wigge, P.A. H2A.Z-containing nucleosomes mediate the thermosensory response in Arabidopsis. Cell 2010, 140, 136–147. [Google Scholar] [CrossRef] [PubMed]
- March-Díaz, R.; Reyes, J.C. The beauty of being a variant: H2A.Z and the SWR1 complex in plants. Mol. Plant 2009, 2, 565–577. [Google Scholar] [CrossRef] [PubMed]
- Zhou, W.; Zhu, Y.; Dong, A.; Shen, W.-H. Histone H2A/H2B chaperones: From molecules to chromatin-based functions in plant growth and development. Plant J. 2015, 83, 78–95. [Google Scholar] [CrossRef] [PubMed]
- Mizuguchi, G. Atp-driven exchange of histone H2AZ variant catalyzed by SWR1 chromatin remodeling complex. Science 2004, 303, 343–348. [Google Scholar] [CrossRef] [PubMed]
- Knizewski, L.; Ginalski, K.; Jerzmanowski, A. Snf2 proteins in plants: Gene silencing and beyond. Trends Plant Sci. 2008, 13, 557–565. [Google Scholar] [CrossRef] [PubMed]
- Jarillo, J.A.; Piñeiro, M. H2A.Z mediates different aspects of chromatin function and modulates flowering responses in Arabidopsis. Plant J. 2015, 83, 96–109. [Google Scholar] [CrossRef] [PubMed]
- Noh, Y.S. Pie1, an iswi family gene, is required for flc activation and floral repression in Arabidopsis. Plant Cell Online 2003, 15, 1671–1682. [Google Scholar] [CrossRef]
- McKinney, E.C.; Kandasamy, M.K.; Meagher, R.B. Arabidopsis contains ancient classes of differentially expressed actin-related protein genes. Plant Physiol. 2002, 128, 997–1007. [Google Scholar] [CrossRef] [PubMed]
- Deal, R.B. The nuclear actin-related protein ARP6 is a pleiotropic developmental regulator required for the maintenance of FLOWERING LOCUS C expression and repression of flowering in Arabidopsis. Plant Cell Online 2005, 17, 2633–2646. [Google Scholar] [CrossRef] [PubMed]
- Kandasamy, M.K.; Deal, R.B.; McKinney, E.C.; Meagher, R.B. Silencing the nuclear actin-related protein atarp4 in Arabidopsis has multiple effects on plant development, including early flowering and delayed floral senescence. Plant J. 2005, 41, 845–858. [Google Scholar] [CrossRef] [PubMed]
- Zacharaki, V.; Benhamed, M.; Poulios, S.; Latrasse, D.; Papoutsoglou, P.; Delarue, M.; Vlachonasios, K.E. The Arabidopsis ortholog of the YEATS domain containing protein YAF9a regulates flowering by controlling H4 acetylation levels at the FLC locus. Plant Sci. 2012, 196, 44–52. [Google Scholar] [CrossRef] [PubMed]
- March-Diaz, R.; Garcia-Dominguez, M.; Florencio, F.J.; Reyes, J.C. SEF, a new protein required for flowering repression in Arabidopsis, interacts with PIE1 and ARP6. Plant Physiol. 2006, 143, 893–901. [Google Scholar] [CrossRef] [PubMed]
- Gómez-Zambrano, Á.; Crevillén, P.; Franco-Zorrilla, J.M.; López, J.A.; Moreno-Romero, J.; Roszak, P.; Santos-González, J.; Jurado, S.; Vázquez, J.; Köhler, C.; et al. Arabidopsis SWC4 binds DNA and recruits the SWR1 complex to modulate histone H2A.Z deposition at key regulatory genes. Mol. Plant 2018, 11, 815–832. [Google Scholar] [CrossRef] [PubMed]
- Cortijo, S.; Charoensawan, V.; Brestovitsky, A.; Buning, R.; Ravarani, C.; Rhodes, D.; van Noort, J.; Jaeger, K.E.; Wigge, P.A. Transcriptional regulation of the ambient temperature response by H2A.Z nucleosomes and HSF1 transcription factors in Arabidopsis. Mol. Plant 2017, 10, 1258–1273. [Google Scholar] [CrossRef] [PubMed]
- Coleman-Derr, D.; Zilberman, D. Deposition of histone variant H2A.Z within gene bodies regulates responsive genes. PLoS Genet. 2012, 8, e1002988. [Google Scholar] [CrossRef] [PubMed]
- Deal, R.B.; Topp, C.N.; McKinney, E.C.; Meagher, R.B. Repression of flowering in Arabidopsis requires activation of flowering locus c expression by the histone variant H2A.Z. Plant Cell Online 2007, 19, 74–83. [Google Scholar] [CrossRef] [PubMed]
- Choi, K.; Kim, S.; Kim, S.Y.; Kim, M.; Hyun, Y.; Lee, H.; Choe, S.; Kim, S.G.; Michaels, S.; Lee, I. Suppressor of frigida3 encodes a nuclear actin-related protein6 required for floral repression in Arabidopsis. Plant Cell 2005, 17, 2647–2660. [Google Scholar] [CrossRef] [PubMed]
- Choi, K.; Park, C.; Lee, J.; Oh, M.; Noh, B.; Lee, I. Arabidopsis homologs of components of the SWR1 complex regulate flowering and plant development. Development 2007, 134, 1931–1941. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sura, W.; Kabza, M.; Karlowski, W.M.; Bieluszewski, T.; Kus-Slowinska, M.; Paweloszek, L.; Sadowski, J.; Ziolkowski, P.A. Dual role of the histone variant H2A.Z in transcriptional regulation of stress-response genes. Plant Cell 2017, 29, 791–807. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gan, E.-S.; Xu, Y.; Wong, J.-Y.; Geraldine Goh, J.; Sun, B.; Wee, W.-Y.; Huang, J.; Ito, T. Jumonji demethylases moderate precocious flowering at elevated temperature via regulation of FLC in Arabidopsis. Nat. Commun. 2014, 5, 5098. [Google Scholar] [CrossRef] [PubMed]
- Gu, X.; Le, C.; Wang, Y.; Li, Z.; Jiang, D.; Wang, Y.; He, Y. Arabidopsis flc clade members form flowering-repressor complexes coordinating responses to endogenous and environmental cues. Nat. Commun. 2013, 4, 1947. [Google Scholar] [CrossRef] [PubMed]
- Fernández, V.; Takahashi, Y.; Le Gourrierec, J.; Coupland, G. Photoperiodic and thermosensory pathways interact through constans to promote flowering at high temperature under short days. Plant J. 2016, 86, 426–440. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.V.; Lucyshyn, D.; Jaeger, K.E.; Alós, E.; Alvey, E.; Harberd, N.P.; Wigge, P.A. Transcription factor PIF4 controls the thermosensory activation of flowering. Nature 2012, 484, 242–245. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, M.; Leichty, A.R.; Hu, T.; Poethig, R.S. H2A.Z promotes the transcription of MIR156A and MIR156C in Arabidopsis by facilitating the deposition of H3K4me3. Development 2018, 145, dev152868. [Google Scholar] [CrossRef] [PubMed]
- Choi, K.; Kim, J.; Müller, S.Y.; Oh, M.; Underwood, C.; Henderson, I.; Lee, I. Regulation of microRNA-mediated developmental changes by the SWR1 chromatin remodeling complex in Arabidopsis thaliana. Plant Physiol. 2016. [Google Scholar] [CrossRef] [PubMed]
- Steffen, A.; Staiger, D. Chromatin marks and ambient temperature-dependent flowering strike up a novel liaison. Genome Boil. 2017, 18, 119. [Google Scholar] [CrossRef] [PubMed]
- Pajoro, A.; Severing, E.; Angenent, G.C.; Immink, R.G.H. Histone H3 lysine 36 methylation affects temperature-induced alternative splicing and flowering in plants. Genome Boil. 2017, 18, 102. [Google Scholar] [CrossRef] [PubMed]
- Luco, R.F.; Pan, Q.; Tominaga, K.; Blencowe, B.J.; Pereira-Smith, O.M.; Misteli, T. Regulation of alternative splicing by histone modifications. Science 2010, 327, 996–1000. [Google Scholar] [CrossRef] [PubMed]
- Bu, Z.; Yu, Y.; Li, Z.; Liu, Y.; Jiang, W.; Huang, Y.; Dong, A.-W. Regulation of Arabidopsis flowering by the histone mark readers MRG1/2 via interaction with constans to modulate FT expression. PLoS Genet. 2014, 10, e1004617. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Gan, E.-S.; Zhou, J.; Wee, W.-Y.; Zhang, X.; Ito, T. Arabidopsis mrg domain proteins bridge two histone modifications to elevate expression of flowering genes. Nucleic Acids Res. 2014, 42, 10960–10974. [Google Scholar] [CrossRef] [PubMed]
- Sureshkumar, S.; Dent, C.; Seleznev, A.; Tasset, C.; Balasubramanian, S. Nonsense-mediated mRNA decay modulates FLM-dependent thermosensory flowering response in Arabidopsis. Nat. Plants 2016, 2, 16055. [Google Scholar] [CrossRef] [PubMed]
- Dubin, M.J.; Zhang, P.; Meng, D.; Remigereau, M.-S.; Osborne, E.J.; Paolo Casale, F.; Drewe, P.; Kahles, A.; Jean, G.; Vilhjálmsson, B.; et al. DNA methylation in Arabidopsis has a genetic basis and shows evidence of local adaptation. eLife 2015, 4, e05255. [Google Scholar] [CrossRef] [PubMed]
- Kawakatsu, T.; Huang, S.-s.C.; Jupe, F.; Sasaki, E.; Schmitz, R.J.; Urich, M.A.; Castanon, R.; Nery, J.R.; Barragan, C.; He, Y.; et al. Epigenomic diversity in a global collection of Arabidopsis thaliana accessions. Cell 2016, 166, 492–505. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Yazaki, J.; Sundaresan, A.; Cokus, S.; Chan, S.W.L.; Chen, H.; Henderson, I.R.; Shinn, P.; Pellegrini, M.; Jacobsen, S.E.; et al. Genome-wide high-resolution mapping and functional analysis of DNA methylation in Arabidopsis. Cell 2006, 126, 1189–1201. [Google Scholar] [CrossRef] [PubMed]
- Stroud, H.; Do, T.; Du, J.; Zhong, X.; Feng, S.; Johnson, L.; Patel, D.J.; Jacobsen, S.E. Non-CG methylation patterns shape the epigenetic landscape in Arabidopsis. Nat. Struct. Mol. Boil. 2013, 21, 64–72. [Google Scholar] [CrossRef] [PubMed]
- Bewick, A.J.; Ji, L.; Niederhuth, C.E.; Willing, E.-M.; Hofmeister, B.T.; Shi, X.; Wang, L.; Lu, Z.; Rohr, N.A.; Hartwig, B.; et al. On the origin and evolutionary consequences of gene body DNA methylation. Proc. Natl. Acad. Sci. USA 2016, 113, 9111–9116. [Google Scholar] [CrossRef] [PubMed]
- Zemach, A.; McDaniel, I.E.; Silva, P.; Zilberman, D. Genome-wide evolutionary analysis of eukaryotic DNA methylation. Science 2010, 328, 916–919. [Google Scholar] [CrossRef] [PubMed]
- Kato, M.; Miura, A.; Bender, J.; Jacobsen, S.E.; Kakutani, T. Role of CG and non-CG methylation in immobilization of transposons in Arabidopsis. Curr. Boil. 2003, 13, 421–426. [Google Scholar] [CrossRef]
- Zhang, H.; Lang, Z.; Zhu, J.-K. Dynamics and function of DNA methylation in plants. Nat. Rev. Mol. Cell Boil. 2018, 19, 489–506. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Harris, C.J.; Liu, Q.; Liu, W.; Ausin, I.; Long, Y.; Xiao, L.; Feng, L.; Chen, X.; Xie, Y.; et al. Large-scale comparative epigenomics reveals hierarchical regulation of non-CG methylation in Arabidopsis. Proc. Natl. Acad. Sci. USA 2018, 115, E1069–E1074. [Google Scholar] [CrossRef] [PubMed]
- Shen, X.; De Jonge, J.; Forsberg, S.K.G.; Pettersson, M.E.; Sheng, Z.; Hennig, L.; Carlborg, Ö. Natural CMT2 variation is associated with genome-wide methylation changes and temperature seasonality. PLoS Genet. 2014, 10, e1004842. [Google Scholar] [CrossRef] [PubMed]
- Jones, A.L.; Sung, S. Mechanisms underlying epigenetic regulation in Arabidopsis thaliana. Integr. Comp. Boil. 2014, 54, 61–67. [Google Scholar] [CrossRef] [PubMed]
- Kinoshita, T.; Miura, A.; Choi, Y.; Kinoshita, Y.; Cao, X.; Jacobsen, S.E.; Fischer, R.L.; Kakutani, T. One-way control of FWA imprinting in Arabidopsis endosperm by DNA methylation. Science 2004, 303, 521–523. [Google Scholar] [CrossRef] [PubMed]
- Soppe, W.J.; Jacobsen, S.E.; Alonso-Blanco, C.; Jackson, J.P.; Kakutani, T.; Koornneef, M.; Peeters, A.J. The late flowering phenotype of fwa mutants is caused by gain-of-function epigenetic alleles of a homeodomain gene. Mol. Cell 2000, 6, 791–802. [Google Scholar] [CrossRef]
- Ikeda, Y.; Kobayashi, Y.; Yamaguchi, A.; Abe, M.; Araki, T. Molecular basis of late-flowering phenotype caused by dominant Epi-alleles of the FWA locus in Arabidopsis. Plant Cell Physiol. 2007, 48, 205–220. [Google Scholar] [CrossRef] [PubMed]
- Yang, R.; Zheng, Z.; Chen, Q.; Yang, L.; Huang, H.; Miki, D.; Wu, W.; Zeng, L.; Liu, J.; Zhou, J.-X.; et al. The developmental regulator PKL is required to maintain correct DNA methylation patterns at RNA-directed DNA methylation loci. Genome Boil. 2017, 18, 103. [Google Scholar] [CrossRef] [PubMed]
- Fu, X.; Li, C.; Liang, Q.; Zhou, Y.; He, H.; Fan, L.-M. CHD3 chromatin-remodeling factor PICKLE regulates floral transition partially via modulating LEAFY expression at the chromatin level in Arabidopsis. Sci. China Life Sci. 2016, 59, 516–528. [Google Scholar] [CrossRef] [PubMed]
- Zha, P.; Jing, Y.; Xu, G.; Lin, R. Pickle chromatin-remodeling factor controls thermosensory hypocotyl growth of Arabidopsis. Plant Cell Environ. 2017, 40, 2426–2436. [Google Scholar] [CrossRef] [PubMed]
- Jing, Y.; Zhang, D.; Wang, X.; Tang, W.; Wang, W.; Huai, J.; Xu, G.; Chen, D.; Li, Y.; Lin, R. Arabidopsis chromatin remodeling factor PICKLE interacts with transcription factor HY5 to regulate hypocotyl cell elongation. Plant Cell 2013, 25, 242–256. [Google Scholar] [CrossRef] [PubMed]
- Marquardt, S.; Raitskin, O.; Wu, Z.; Liu, F.; Sun, Q.; Dean, C. Functional consequences of splicing of the antisense transcript coolair on flc transcription. Mol. Cell 2014, 54, 156–165. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Swiezewski, S.; Liu, F.; Magusin, A.; Dean, C. Cold-induced silencing by long antisense transcripts of an Arabidopsis polycomb target. Nature 2009, 462, 799–802. [Google Scholar] [CrossRef] [PubMed]
- Csorba, T.; Questa, J.I.; Sun, Q.; Dean, C. Antisense coolair mediates the coordinated switching of chromatin states at flc during vernalization. Proc. Natl. Acad. Sci. USA 2014, 111, 16160–16165. [Google Scholar] [CrossRef] [PubMed]
- Kim, W.; Ahn, H.J.; Chiou, T.-J.; Ahn, J.H. The role of the miR399-PHO2 module in the regulation of flowering time in response to different ambient temperatures in Arabidopsis thaliana. Mol. Cells 2011, 32, 83–88. [Google Scholar] [CrossRef] [PubMed]
- Quinn, J.J.; Chang, H.Y. Unique features of long non-coding RNA biogenesis and function. Nat. Rev. Genet. 2016, 17, 47. [Google Scholar] [CrossRef] [PubMed]
- Kung, J.T.; Colognori, D.; Lee, J.T. Long noncoding RNAs: Past, present, and future. Genetics 2013, 193, 651–669. [Google Scholar] [CrossRef] [PubMed]
- Kornienko, A.E.; Guenzl, P.M.; Barlow, D.P.; Pauler, F.M. Gene regulation by the act of long non-coding RNA transcription. BMC Boil. 2013, 11, 59. [Google Scholar] [CrossRef] [PubMed]
- Geisler, S.; Coller, J. RNA in unexpected places: Long non-coding RNA functions in diverse cellular contexts. Nat. Rev. Mol. Cell Boil. 2013, 14, 699. [Google Scholar] [CrossRef] [PubMed]
- Heo, J.B.; Sung, S. Vernalization-mediated epigenetic silencing by a long intronic noncoding RNA. Science 2011, 331, 76–79. [Google Scholar] [CrossRef] [PubMed]
- Bäurle, I.; Dean, C. Differential interactions of the autonomous pathway RRM proteins and chromatin regulators in the silencing of Arabidopsis targets. PLoS ONE 2008, 3, e2733. [Google Scholar] [CrossRef] [PubMed]
- Hornyik, C.; Terzi, L.C.; Simpson, G.G. The spen family protein FPA controls alternative cleavage and polyadenylation of RNA. Dev. Cell 2010, 18, 203–213. [Google Scholar] [CrossRef] [PubMed]
- Rosonina, E.; Manley, J.L. Alternative polyadenylation blooms. Dev. Cell 2010, 18, 172–174. [Google Scholar] [CrossRef] [PubMed]
- Severing, E.; Faino, L.; Jamge, S.; Busscher, M.; Kuijer-Zhang, Y.; Bellinazzo, F.; Busscher-Lange, J.; Fernández, V.; Angenent, G.C.; Immink, R.G. Arabidopsis thaliana ambient temperature responsive lncRNAs. BMC Plant Boil. 2018, 18, 145. [Google Scholar] [CrossRef] [PubMed]
- Gyula, P.; Baksa, I.; Tóth, T.; Mohorianu, I.; Dalmay, T.; Szittya, G. Ambient temperature regulates the expression of a small set of sRNAs influencing plant development through NF-YA2 and YUC2. Plant Cell Environ. 2018. [Google Scholar] [CrossRef] [PubMed]
- Bartel, D.P. Micrornas: Genomics, biogenesis, mechanism, and function. Cell 2004, 116, 281–297. [Google Scholar] [CrossRef]
- Chen, X. A microRNA as a translational repressor of apetala2 in Arabidopsis flower development. Science 2004, 303, 2022–2025. [Google Scholar] [CrossRef] [PubMed]
- Johnson, C.R.; Millwood, R.J.; Tang, Y.; Gou, J.; Sykes, R.W.; Turner, G.B.; Davis, M.F.; Sang, Y.; Wang, Z.-Y.; Stewart, C.N. Field-grown miR156 transgenic switchgrass reproduction, yield, global gene expression analysis, and bioconfinement. Biotechnol. Biofuels 2017, 10, 255. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sunkar, R.; Li, Y.-F.; Jagadeeswaran, G. Functions of microRNAs in plant stress responses. Trends Plant Sci. 2012, 17, 196–203. [Google Scholar] [CrossRef] [PubMed]
- Hu, J.-Y.; Zhou, Y.; He, F.; Dong, X.; Liu, L.-Y.; Coupland, G.; Turck, F.; de Meaux, J. miR824-regulated AGAMOUS-LIKE16 contributes to flowering time repression in Arabidopsis. Plant Cell 2014, 26, 2024–2037. [Google Scholar] [CrossRef] [PubMed]
- Huo, H.; Wei, S.; Bradford, K.J. Delay of GERMINATION1 (DOG1) regulates both seed dormancy and flowering time through microRNA pathways. Proc. Natl. Acad. Sci. USA 2016, 113, E2199–E2206. [Google Scholar] [CrossRef] [PubMed]
- Teotia, S.; Tang, G. To bloom or not to bloom: Role of microRNAs in plant flowering. Mol. Plant 2015, 8, 359–377. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.; Yoo, S.J.; Lee, J.H.; Kim, W.; Yoo, S.K.; Fitzgerald, H.; Carrington, J.C.; Ahn, J.H. Genetic framework for flowering-time regulation by ambient temperature-responsive mirnas in Arabidopsis. Nucleic Acids Res. 2010, 38, 3081–3093. [Google Scholar] [CrossRef] [PubMed]
- Poethig, R.S. Small RNAs and developmental timing in plants. Curr. Opin. Genet. Dev. 2009, 19, 374–378. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chuck, G.; Cigan, A.M.; Saeteurn, K.; Hake, S. The heterochronic maize mutant Corngrass1 results from overexpression of a tandem microRNA. Nat. Genet. 2007, 39, 544. [Google Scholar] [CrossRef] [PubMed]
- Preston, J.C.; Hileman, L. Functional evolution in the plant SQUAMOSA-PROMOTER BINDING PROTEIN-LIKE (SPL) gene family. Front. Plant Sci. 2013, 4, 80. [Google Scholar] [CrossRef] [PubMed]
- Srikanth, A.; Schmid, M. Regulation of flowering time: All roads lead to rome. Cell. Mol. Life Sci. 2011, 68, 2013–2037. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.-W.; Czech, B.; Weigel, D. miR156-regulated SPL transcription factors define an endogenous flowering pathway in Arabidopsis thaliana. Cell 2009, 138, 738–749. [Google Scholar] [CrossRef] [PubMed]
- Yamaguchi, A.; Wu, M.-F.; Yang, L.; Wu, G.; Poethig, R.S.; Wagner, D. The microRNA-regulated SBP-Box transcription factor SPL3 is a direct upstream activator of leafy, fruitfull, and apetala1. Dev. Cell 2009, 17, 268–278. [Google Scholar] [CrossRef] [PubMed]
- Wigge, P.A.; Kim, M.C.; Jaeger, K.E.; Busch, W.; Schmid, M.; Lohmann, J.U.; Weigel, D. Integration of spatial and temporal information during floral induction in Arabidopsis. Science 2005, 309, 1056–1059. [Google Scholar] [CrossRef] [PubMed]
- Abe, M.; Kobayashi, Y.; Yamamoto, S.; Daimon, Y.; Yamaguchi, A.; Ikeda, Y.; Ichinoki, H.; Notaguchi, M.; Goto, K.; Araki, T. Fd, a bZIP protein mediating signals from the floral pathway integrator FT at the shoot apex. Science 2005, 309, 1052–1056. [Google Scholar] [CrossRef] [PubMed]
- Huijser, P.; Schmid, M. The control of developmental phase transitions in plants. Development 2011, 138, 4117–4129. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zheng, Q.; Liu, J.; Goff, B.M.; Dinkins, R.D.; Zhu, H. Genetic manipulation of miR156 for improvement of biomass production and forage quality in red clover. Crop. Sci. 2016, 56, 1199–1205. [Google Scholar] [CrossRef]
- Fu, C.; Sunkar, R.; Zhou, C.; Shen, H.; Zhang, J.Y.; Matts, J.; Wolf, J.; Mann, D.G.; Stewart, C.N.; Tang, Y. Overexpression of miR156 in switchgrass (Panicum virgatum L.) results in various morphological alterations and leads to improved biomass production. Plant Biotechnol. J. 2012, 10, 443–452. [Google Scholar] [CrossRef] [PubMed]
- Wu, Z.; Cao, Y.; Yang, R.; Qi, T.; Hang, Y.; Lin, H.; Zhou, G.; Wang, Z.-Y.; Fu, C. Switchgrass SBP-Box transcription factors PvSPL1 and 2 function redundantly to initiate side tillers and affect biomass yield of energy crop. Biotechnol. Biofuels 2016, 9, 101. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Zou, Z.; Zhang, J.; Zhang, Y.; Han, Q.; Hu, T.; Xu, X.; Liu, H.; Li, H.; Ye, Z. Over-expression of sly-miR156a in tomato results in multiple vegetative and reproductive trait alterations and partial phenocopy of the sft mutant. FEBS Lett. 2011, 585, 435–439. [Google Scholar] [CrossRef] [PubMed]
- Cho, H.J.; Kim, J.J.; Lee, J.H.; Kim, W.; Jung, J.-H.; Park, C.-M.; Ahn, J.H. SHORT VEGETATIVE PHASE (SVP) protein negatively regulates miR172 transcription via direct binding to the pri-miR172a promoter in Arabidopsis. FEBS Lett. 2012, 586, 2332–2337. [Google Scholar] [CrossRef] [PubMed]
- Wu, G.; Park, M.Y.; Conway, S.R.; Wang, J.-W.; Weigel, D.; Poethig, R.S. The sequential action of miR156 and miR172 regulates developmental timing in Arabidopsis. Cell 2009, 138, 750–759. [Google Scholar] [CrossRef] [PubMed]
- Aukerman, M.J.; Sakai, H. Regulation of flowering time and floral organ identity by a microRNA and its apetala2-like target genes. Plant Cell 2003, 15, 2730–2741. [Google Scholar] [CrossRef] [PubMed]
- Jung, J.-H.; Seo, Y.-H.; Seo, P.J.; Reyes, J.L.; Yun, J.; Chua, N.-H.; Park, C.-M. The gigantea-regulated microRNA172 mediates photoperiodic flowering independent of constans in Arabidopsis. Plant Cell 2007, 19, 2736–2748. [Google Scholar] [CrossRef] [PubMed]
- Mathieu, J.; Yant, L.J.; Mürdter, F.; Küttner, F.; Schmid, M. Repression of flowering by the miR172 target SMZ. PLoS Boil. 2009, 7, e1000148. [Google Scholar] [CrossRef] [PubMed]
- Schmid, M.; Uhlenhaut, N.H.; Godard, F.; Demar, M.; Bressan, R.; Weigel, D.; Lohmann, J.U. Dissection of floral induction pathways using global expression analysis. Development 2003, 130, 6001–6012. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, Y.-S.; Lee, D.-Y.; Cho, L.-H.; An, G. Rice miR172 induces flowering by suppressing OsiDS1 and SNB, two AP2 genes that negatively regulate expression of Ehd1 and florigens. Rice 2014, 7, 31. [Google Scholar] [CrossRef] [PubMed]
- Lauter, N.; Kampani, A.; Carlson, S.; Goebel, M.; Moose, S.P. MicroRNA172 down-regulates glossy15 to promote vegetative phase change in maize. Proc. Natl. Acad. Sci. USA 2005, 102, 9412–9417. [Google Scholar] [CrossRef] [PubMed]
- Cho, S.H.; Coruh, C.; Axtell, M.J. MiR156 and miR390 regulate tasirna accumulation and developmental timing in physcomitrella patens. Plant Cell 2012, 24, 4837–4849. [Google Scholar] [CrossRef] [PubMed]
- Tao, Z.; Shen, L.; Liu, C.; Liu, L.; Yan, Y.; Yu, H. Genome-wide identification of SOC1 and SVP targets during the floral transition in Arabidopsis. Plant J. 2012, 70, 549–561. [Google Scholar] [CrossRef] [PubMed]
- Posé, D.; Verhage, L.; Ott, F.; Yant, L.; Mathieu, J.; Angenent, G.C.; Immink, R.G.; Schmid, M. Temperature-dependent regulation of flowering by antagonistic FLM variants. Nature 2013, 503, 414–417. [Google Scholar] [CrossRef] [PubMed]
- Kim, W.; Kim, H.-E.; Jun, A.R.; Jung, M.G.; Jin, S.; Lee, J.-H.; Ahn, J.H. Structural determinants of miR156a precursor processing in temperature-responsive flowering in Arabidopsis. J. Exp. Bot. 2016, 67, 4659–4670. [Google Scholar] [CrossRef] [PubMed]
- Cuperus, J.T.; Fahlgren, N.; Carrington, J.C. Evolution and functional diversification ofmirnagenes. Plant Cell 2011, 23, 431–442. [Google Scholar] [CrossRef] [PubMed]
- Bologna, N.G.; Schapire, A.L.; Zhai, J.; Chorostecki, U.; Boisbouvier, J.; Meyers, B.C.; Palatnik, J.F. Multiple RNA recognition patterns during microRNA biogenesis in plants. Genome Res. 2013, 23, 1675–1689. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bologna, N.G.; Mateos, J.L.; Bresso, E.G.; Palatnik, J.F. A loop-to-base processing mechanism underlies the biogenesis of plant micrornas miR319 and miR159. EMBO J. 2009, 28, 3646–3656. [Google Scholar] [CrossRef] [PubMed]
- Mateos, J.L.; Bologna, N.G.; Chorostecki, U.; Palatnik, J.F. Identification of microrna processing determinants by random mutagenesis of Arabidopsis miR172a precursor. Curr. Boil. 2010, 20, 49–54. [Google Scholar] [CrossRef] [PubMed]
- Song, L.; Axtell, M.J.; Fedoroff, N.V. Rna secondary structural determinants of miRNA precursor processing in Arabidopsis. Curr. Boil. 2010, 20, 37–41. [Google Scholar] [CrossRef] [PubMed]
- Werner, S.; Wollmann, H.; Schneeberger, K.; Weigel, D. Structure determinants for accurate processing of miR172a in Arabidopsis thaliana. Curr. Boil. 2010, 20, 42–48. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.-E.; Kim, W.; Lee, A.-R.; Jin, S.; Jun, A.R.; Kim, N.-K.; Lee, J.-H.; Ahn, J.H. Base-pair opening dynamics of the microRNA precursor pri-miR156a affect temperature-responsive flowering in Arabidopsis. Biochem. Biophys. Res. Commun. 2017, 484, 839–844. [Google Scholar] [CrossRef] [PubMed]
- Kim, W.; Kim, H.-E.; Lee, A.-R.; Jun, A.R.; Jung, M.G.; Ahn, J.H.; Lee, J.-H. Base-pair opening dynamics of primary miR156a using NMR elucidates structural determinants important for its processing level and leaf number phenotype in Arabidopsis. Nucleic Acids Res. 2017, 45, 875–885. [Google Scholar] [CrossRef] [PubMed]
- Jung, J.-H.; Seo, P.J.; Ahn, J.-H.; Park, C.-M. The Arabidopsis RNA-binding protein FCA regulates microRNA172 processing in thermosensory flowering. J. Boil. Chem. 2012, 287, 16007–16016. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.H.; Ryu, H.-S.; Chung, K.S.; Posé, D.; Kim, S.; Schmid, M.; Ahn, J.H. Regulation of temperature-responsive flowering by mads-box transcription factor repressors. Science 2013, 342, 628–632. [Google Scholar] [CrossRef] [PubMed]
- Theissen, G.; Becker, A.; Di Rosa, A.; Kanno, A.; Kim, J.T.; Münster, T.; Winter, K.-U.; Saedler, H. A short history of mads-box genes in plants. In Plant Molecular Evolution; Springer: Dordrecht, The Netherlands, 2000; pp. 115–149. [Google Scholar]
- Theißen, G. Development of floral organ identity: Stories from the mads house. Curr. Opin. Plant Boil. 2001, 4, 75–85. [Google Scholar] [CrossRef]
- Ng, M.; Yanofsky, M.F. Function and evolution of the plant mads-box gene family. Nat. Rev. Genet. 2001, 2, 186. [Google Scholar] [CrossRef] [PubMed]
- Ratcliffe, O.J.; Nadzan, G.C.; Reuber, T.L.; Riechmann, J.L. Regulation of flowering in Arabidopsis by an FLC homologue. Plant Physiol. 2001, 126, 122–132. [Google Scholar] [CrossRef] [PubMed]
- Smaczniak, C.; Immink, R.G.; Angenent, G.C.; Kaufmann, K. Developmental and evolutionary diversity of plant mads-domain factors: Insights from recent studies. Development 2012, 139, 3081–3098. [Google Scholar] [CrossRef] [PubMed]
- Balasubramanian, S.; Weigel, D. Temperature induced flowering in Arabidopsis thaliana. Plant Signal. Behav. 2006, 1, 227–228. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.H.; Yoo, S.J.; Park, S.H.; Hwang, I.; Lee, J.S.; Ahn, J.H. Role of SVP in the control of flowering time by ambient temperature in Arabidopsis. Genes Dev. 2007, 21, 397–402. [Google Scholar] [CrossRef] [PubMed]
- Werner, J.D.; Borevitz, J.O.; Warthmann, N.; Trainer, G.T.; Ecker, J.R.; Chory, J.; Weigel, D. Quantitative trait locus mapping and DNA array hybridization identify an FLM deletion as a cause for natural flowering-time variation. Proc. Natl. Acad. Sci. USA 2005, 102, 2460–2465. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lutz, U.; Posé, D.; Pfeifer, M.; Gundlach, H.; Hagmann, J.; Wang, C.; Weigel, D.; Mayer, K.F.; Schmid, M.; Schwechheimer, C. Modulation of ambient temperature-dependent flowering in Arabidopsis thaliana by natural variation of FLOWERING LOCUS M. PLoS Genet. 2015, 11, e1005588. [Google Scholar] [CrossRef] [PubMed]
- Méndez-Vigo, B.; Martínez-Zapater, J.M.; Alonso-Blanco, C. The flowering repressor SVP underlies a novel Arabidopsis thaliana QTL interacting with the genetic background. PLoS Genet. 2013, 9, e1003289. [Google Scholar] [CrossRef] [PubMed]
- Scortecci, K.; Michaels, S.D.; Amasino, R.M. Genetic interactions between FLM and other flowering-time genes in Arabidopsis thaliana. Plant Mol. Boil. 2003, 52, 915–922. [Google Scholar] [CrossRef]
- Capovilla, G.; Symeonidi, E.; Wu, R.; Schmid, M. Contribution of major FLM isoforms to temperature-dependent flowering in Arabidopsis thaliana. J. Exp. Bot. 2017, 68, 5117–5127. [Google Scholar] [CrossRef] [PubMed]
- Lee, K.C.; Jang, Y.H.; Kim, S.-K.; Park, H.-Y.; Thu, M.P.; Lee, J.H.; Kim, J.-K. Rrm domain of Arabidopsis splicing factor SF1 is important for pre-mRNA splicing of a specific set of genes. Plant Cell Rep. 2017, 36, 1083–1095. [Google Scholar] [CrossRef] [PubMed]
- Drechsel, G.; Kahles, A.; Kesarwani, A.K.; Stauffer, E.; Behr, J.; Drewe, P.; Rätsch, G.; Wachter, A. Nonsense-mediated decay of alternative precursor mRNA splicing variants is a major determinant of the Arabidopsis steady state transcriptome. Plant Cell 2013, 25, 3726–3742. [Google Scholar] [CrossRef] [PubMed]
- Ye, C.Y.; Chen, L.; Liu, C.; Zhu, Q.H.; Fan, L. Widespread noncoding circular RNAs in plants. New Phytol. 2015, 208, 88–95. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Conn, V.M.; Hugouvieux, V.; Nayak, A.; Conos, S.A.; Capovilla, G.; Cildir, G.; Jourdain, A.; Tergaonkar, V.; Schmid, M.; Zubieta, C. A circrna from sepallata3 regulates splicing of its cognate mrna through r-loop formation. Nat. Plants 2017, 3, 17053. [Google Scholar] [CrossRef] [PubMed]
- Cheng, J.; Zhang, Y.; Li, Z.; Wang, T.; Zhang, X.; Zheng, B. A lariat-derived circular RNA is required for plant development in Arabidopsis. Sci. China Life Sci. 2018, 61, 204–213. [Google Scholar] [CrossRef] [PubMed]
- Ratcliffe, O.J.; Kumimoto, R.W.; Wong, B.J.; Riechmann, J.L. Analysis of the Arabidopsis mads affecting flowering gene family: Maf2 prevents vernalization by short periods of cold. Plant Cell 2003, 15, 1159–1169. [Google Scholar] [CrossRef] [PubMed]
- Rosloski, S.M.; Singh, A.; Jali, S.S.; Balasubramanian, S.; Weigel, D.; Grbic, V. Functional analysis of splice variant expression of mads affecting flowering 2 of Arabidopsis thaliana. Plant Mol. Boil. 2013, 81, 57–69. [Google Scholar] [CrossRef] [PubMed]
- Salomé, P.A.; Bomblies, K.; Laitinen, R.A.; Yant, L.; Mott, R.; Weigel, D. Genetic architecture of flowering time variation in Arabidopsis thaliana. Genetics 2011, 188, 421–433. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Airoldi, C.A.; McKay, M.; Davies, B. MAF2 is regulated by temperature-dependent splicing and represses flowering at low temperatures in parallel with FLM. PLoS ONE 2015, 10, e0126516. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Jack, T. Defining subdomains of the k domain important for protein–protein interactions of plant mads proteins. Plant Mol. Boil. 2004, 55, 45–59. [Google Scholar] [CrossRef] [PubMed]
- Ma, H.; Yanofsky, M.F.; Meyerowitz, E.M. AGL1-AGL6, an Arabidopsis gene family with similarity to floral homeotic and transcription factor genes. Genes Dev. 1991, 5, 484–495. [Google Scholar] [CrossRef] [PubMed]
- Hu, X.; Kong, X.; Wang, C.; Ma, L.; Zhao, J.; Wei, J.; Zhang, X.; Loake, G.J.; Zhang, T.; Huang, J. Proteasome-mediated degradation of FRIGIDA modulates flowering time in Arabidopsis during vernalization. Plant Cell Online 2014, 26, 4763–4781. [Google Scholar] [CrossRef] [PubMed]
- Johanson, U.; West, J.; Lister, C.; Michaels, S.; Amasino, R.; Dean, C. Molecular analysis of frigida, a major determinant of natural variation in Arabidopsis flowering time. Science 2000, 290, 344–347. [Google Scholar] [CrossRef] [PubMed]
- Hepworth, S.R.; Valverde, F.; Ravenscroft, D.; Mouradov, A.; Coupland, G. Antagonistic regulation of flowering-time gene SOC1 by constans and FLC via separate promoter motifs. EMBO J. 2002, 21, 4327–4337. [Google Scholar] [CrossRef] [PubMed]
- Michaels, S.D.; Himelblau, E.; Kim, S.Y.; Schomburg, F.M.; Amasino, R.M. Integration of flowering signals in winter-annual Arabidopsis. Plant Physiol. 2005, 137, 149–156. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.-H.; Doyle, M.R.; Sung, S.; Amasino, R.M. Vernalization: Winter and the timing of flowering in plants. Annu. Rev. Cell Dev. 2009, 25, 277–299. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.-J.; Zhang, Y.-C.; Li, Q.-H.; Sang, Y.; Mao, J.; Lian, H.-L.; Wang, L.; Yang, H.-Q. COP1-mediated ubiquitination of CONSTANS is implicated in cryptochrome regulation of flowering in Arabidopsis. Plant Cell 2008, 20, 292–306. [Google Scholar] [CrossRef] [PubMed]
- Yu, J.-W.; Rubio, V.; Lee, N.-Y.; Bai, S.; Lee, S.-Y.; Kim, S.-S.; Liu, L.; Zhang, Y.; Irigoyen, M.L.; Sullivan, J.A.; et al. COP1 and ELF3 control circadian function and photoperiodic flowering by regulating GI stability. Mol. Cell 2008, 32, 617–630. [Google Scholar] [CrossRef] [PubMed]
- Jang, K.; Lee, H.G.; Jung, S.-J.; Paek, N.-C.; Seo, P.J. The E3 ubiquitin ligase COP1 regulates thermosensory flowering by triggering GI degradation in Arabidopsis. Sci. Rep. 2015, 5, 12071. [Google Scholar] [CrossRef] [PubMed]
- Sawa, M.; Kay, S.A. GIGANTEA directly activates Flowering Locus T in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 2011, 108, 11698–11703. [Google Scholar] [CrossRef] [PubMed]
- Gangappa, S.N.; Kumar, S.V. DET1 and HY5 control PIF4-mediated thermosensory elongation growth through distinct mechanisms. Cell Rep. 2017, 18, 344–351. [Google Scholar] [CrossRef] [PubMed]
- Park, Y.-J.; Lee, H.-J.; Ha, J.-H.; Kim, J.Y.; Park, C.-M. COP1 conveys warm temperature information to hypocotyl thermomorphogenesis. New Phytol. 2017, 215, 269–280. [Google Scholar] [CrossRef] [PubMed]
- Van den Besselaar, E.J.M.; Sanchez-Lorenzo, A.; Wild, M.; Klein Tank, A.M.G.; de Laat, A.T.J. Relationship between sunshine duration and temperature trends across europe since the second half of the twentieth century. J. Geophys. Res. Atmos. 2015, 120, 10823–10836. [Google Scholar] [CrossRef]
- Makowski, K.; Jaeger, E.B.; Chiacchio, M.; Wild, M.; Ewen, T.; Ohmura, A. On the relationship between diurnal temperature range and surface solar radiation in europe. J. Geophys. Res. 2009, 114. [Google Scholar] [CrossRef]
- Ye, J.; Li, F.; Sun, G.; Guo, A. Solar dimming and its impact on estimating solar radiation from diurnal temperature range in China, 1961–2007. Theor. Appl. Clim. 2009, 101, 137–142. [Google Scholar] [CrossRef]
- Legris, M.; Nieto, C.; Sellaro, R.; Prat, S.; Casal, J.J. Perception and signalling of light and temperature cues in plants. Plant J. 2017, 90, 683–697. [Google Scholar] [CrossRef] [PubMed]
- Delker, C.; Sonntag, L.; James, G.V.; Janitza, P.; Ibañez, C.; Ziermann, H.; Peterson, T.; Denk, K.; Mull, S.; Ziegler, J.; et al. The DET1-COP1-HY5 pathway constitutes a multipurpose signaling module regulating plant photomorphogenesis and thermomorphogenesis. Cell Rep. 2014, 9, 1983–1989. [Google Scholar] [CrossRef] [PubMed]
- Jung, J.-H.; Domijan, M.; Klose, C.; Biswas, S.; Ezer, D.; Gao, M.; Khattak, A.K.; Box, M.S.; Charoensawan, V.; Cortijo, S.; et al. Phytochromes function as thermosensors in Arabidopsis. Science 2016, 354, 886–889. [Google Scholar] [CrossRef] [PubMed]
- Heschel, M.S.; Butler, C.M.; Barua, D.; Chiang, G.C.K.; Wheeler, A.; Sharrock, R.A.; Whitelam, G.C.; Donohue, K. New roles of phytochromes during seed germination. Int. J. Plant Sci. 2008, 169, 531–540. [Google Scholar] [CrossRef]
- Dechaine, J.M.; Gardner, G.; Weinig, C. Phytochromes differentially regulate seed germination responses to light quality and temperature cues during seed maturation. Plant Cell Environ. 2009, 32, 1297–1309. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Blazquez, M.A.; Ahn, J.H.; Weigel, D. A thermosensory pathway controlling flowering time in Arabidopsis thaliana. Nat Genet. 2003, 33, 168–171. [Google Scholar] [CrossRef] [PubMed]
- Halliday, K.J.; Salter, M.G.; Thingnaes, E.; Whitelam, G.C. Phytochrome control of flowering is temperature sensitive and correlates with expression of the floral integrator FT. Plant J 2003, 33, 875–885. [Google Scholar] [CrossRef] [PubMed]
- Song, Y.H.; Ito, S.; Imaizumi, T. Flowering time regulation: Photoperiod- and temperature-sensing in leaves. Trends Plant Sci. 2013, 18, 575–583. [Google Scholar] [CrossRef] [PubMed]
- Legris, M.; Klose, C.; Burgie, E.S.; Rojas, C.C.R.; Neme, M.; Hiltbrunner, A.; Wigge, P.A.; Schäfer, E.; Vierstra, R.D.; Casal, J.J. Phytochrome B integrates light and temperature signals in Arabidopsis. Science 2016, 354, 897–900. [Google Scholar] [CrossRef] [PubMed]
- Delker, C.; van Zanten, M.; Quint, M. Thermosensing enlightened. Trends Plant Sci. 2017, 22, 185–187. [Google Scholar] [CrossRef] [PubMed]
- Taylor, A.O. In vitro phytochrome dark reversion process. Plant Physiol. 1968, 43, 767–774. [Google Scholar] [CrossRef] [PubMed]
- Klose, C.; Venezia, F.; Hussong, A.; Kircher, S.; Schäfer, E.; Fleck, C. Systematic analysis of how phytochrome B dimerization determines its specificity. Nat. Plants 2015, 1, 15090. [Google Scholar] [CrossRef] [PubMed]
- Franklin, K.A.; Quail, P.H. Phytochrome functions in Arabidopsis development. J. Exp. Bot. 2009, 61, 11–24. [Google Scholar] [CrossRef] [PubMed]
- Quail, P.H. Phytochrome photosensory signalling networks. Nat. Rev. Mol. Cell Boil. 2002, 3, 85–93. [Google Scholar] [CrossRef] [PubMed]
- Hu, W.; Franklin, K.A.; Sharrock, R.A.; Jones, M.A.; Harmer, S.L.; Lagarias, J.C. Unanticipated regulatory roles forarabidopsisphytochromes revealed by null mutant analysis. Proc. Natl. Acad. Sci. USA 2013, 110, 1542–1547. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Smith, R.W.; Helwig, B.; Westphal, A.H.; Pel, E.; Borst, J.W.; Fleck, C. Interactions between phyB and PIF proteins alter thermal reversion reactions in vitro. Photochem. Photobiol. 2017, 93, 1525–1531. [Google Scholar] [CrossRef] [PubMed]
- Ni, W.; Xu, S.L.; Tepperman, J.M.; Stanley, D.J.; Maltby, D.A.; Gross, J.D.; Burlingame, A.L.; Wang, Z.Y.; Quail, P.H. A mutually assured destruction mechanism attenuates light signaling in Arabidopsis. Science 2014, 344, 1160–1164. [Google Scholar] [CrossRef] [PubMed]
- Valverde, F. Photoreceptor regulation of constans protein in photoperiodic flowering. Science 2004, 303, 1003–1006. [Google Scholar] [CrossRef] [PubMed]
- Lorrain, S.; Allen, T.; Duek, P.D.; Whitelam, G.C.; Fankhauser, C. Phytochrome-mediated inhibition of shade avoidance involves degradation of growth-promoting bhlh transcription factors. Plant J. 2007, 53, 312–323. [Google Scholar] [CrossRef] [PubMed]
- Kaiserli, E.; Páldi, K.; O’Donnell, L.; Batalov, O.; Pedmale, U.V.; Nusinow, D.A.; Kay, S.A.; Chory, J. Integration of light and photoperiodic signaling in transcriptional nuclear foci. Dev. Cell 2015, 35, 311–321. [Google Scholar] [CrossRef] [PubMed]
- Hajdu, A.; Ádám, É.; Sheerin, D.J.; Dobos, O.; Bernula, P.; Hiltbrunner, A.; Kozma-Bognár, L.; Nagy, F. High-level expression and phosphorylation of phytochrome B modulates flowering time in Arabidopsis. Plant J. 2015, 83, 794–805. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fujii, Y.; Tanaka, H.; Konno, N.; Ogasawara, Y.; Hamashima, N.; Tamura, S.; Hasegawa, S.; Hayasaki, Y.; Okajima, K.; Kodama, Y. Phototropin perceives temperature based on the lifetime of its photoactivated state. Proc. Natl. Acad. Sci. USA 2017, 114, 9206–9211. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Inoue, S.-I.; Takemiya, A.; Shimazaki, K.-I. Phototropin signaling and stomatal opening as a model case. Curr. Opin. Plant Boil. 2010, 13, 587–593. [Google Scholar] [CrossRef] [PubMed]
- Takemiya, A. Phototropins promote plant growth in response to blue light in low light environments. Plant Cell Online 2005, 17, 1120–1127. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Yu, X.; Li, K.; Klejnot, J.; Yang, H.; Lisiero, D.; Lin, C. Photoexcited CRY2 interacts with CIB1 to regulate transcription and floral initiation in Arabidopsis. Science 2008, 322, 1535–1539. [Google Scholar] [CrossRef] [PubMed]
- Ma, D.; Li, X.; Guo, Y.; Chu, J.; Fang, S.; Yan, C.; Noel, J.P.; Liu, H. Cryptochrome 1 interacts with pif4 to regulate high temperature-mediated hypocotyl elongation in response to blue light. Proc. Natl. Acad. Sci. USA 2016, 113, 224–229. [Google Scholar] [CrossRef] [PubMed]
- Huang, H.; Nusinow, D.A. Into the evening: Complex interactions in the Arabidopsis circadian clock. Trends Genet. 2016, 32, 674–686. [Google Scholar] [CrossRef] [PubMed]
- Chow, B.Y.; Kay, S.A. Global approaches for telling time: Omics and the Arabidopsis circadian clock. Semin. Cell Dev. Boil. 2013, 24, 383–392. [Google Scholar] [CrossRef] [PubMed]
- McClung, C.R. Plant circadian rhythms. Plant Cell Online 2006, 18, 792–803. [Google Scholar] [CrossRef] [PubMed]
- Hsu, P.Y.; Harmer, S.L. Wheels within wheels: The plant circadian system. Trends Plant Sci. 2014, 19, 240–249. [Google Scholar] [CrossRef] [PubMed]
- Oakenfull, R.J.; Davis, S.J. Shining a light on the Arabidopsis circadian clock. Plant Cell Environ. 2017, 40, 2571–2585. [Google Scholar] [CrossRef] [PubMed]
- Kusakina, J.; Gould, P.D.; Hall, A. A fast circadian clock at high temperatures is a conserved feature acrossarabidopsisaccessions and likely to be important for vegetative yield. Plant Cell Environ. 2014, 37, 327–340. [Google Scholar] [CrossRef] [PubMed]
- Boikoglou, E.; Ma, Z.; von Korff, M.; Davis, A.M.; Nagy, F.; Davis, S.J. Environmental memory from a circadian oscillator: The Arabidopsis thaliana clock differentially integrates perception of photic vs. Thermal entrainment. Genetics 2011, 189, 655–664. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Eckardt, N.A. A wheel within a wheel: Temperature compensation of the circadian clock. Plant Cell Online 2006, 18, 1105–1108. [Google Scholar] [CrossRef]
- Eckardt, N.A. Temperature entrainment of the Arabidopsis circadian clock. Plant Cell Online 2005, 17, 645–647. [Google Scholar] [CrossRef]
- Salome, P.A.; Weigel, D.; McClung, C.R. The role of the Arabidopsis morning loop components CCA1, LHY, PRR7, and PRR9 in temperature compensation. Plant Cell Online 2010, 22, 3650–3661. [Google Scholar] [CrossRef] [PubMed]
- Gould, P.D. The molecular basis of temperature compensation in the Arabidopsis circadian clock. Plant Cell Online 2006, 18, 1177–1187. [Google Scholar] [CrossRef] [PubMed]
- Mizuno, T.; Nomoto, Y.; Oka, H.; Kitayama, M.; Takeuchi, A.; Tsubouchi, M.; Yamashino, T. Ambient temperature signal feeds into the circadian clock transcriptional circuitry through the EC night-time repressor in Arabidopsis thaliana. Plant Cell Physiol. 2014, 55, 958–976. [Google Scholar] [CrossRef] [PubMed]
- Thines, B.; Harmon, F.G. Ambient temperature response establishes ELF3 as a required component of the core Arabidopsis circadian clock. Proc. Natl. Acad. Sci. USA 2010, 107, 3257–3262. [Google Scholar] [CrossRef] [PubMed]
- Box, M.S.; Huang, B.E.; Domijan, M.; Jaeger, K.E.; Khattak, A.K.; Yoo, S.J.; Sedivy, E.L.; Jones, D.M.; Hearn, T.J.; Webb, A.A.R.; et al. ELF3 controls thermoresponsive growth in Arabidopsis. Curr. Boil. 2015, 25, 194–199. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nagel, D.H.; Pruneda-Paz, J.L.; Kay, S.A. FBH1 affects warm temperature responses in the Arabidopsis circadian clock. Proc. Natl. Acad. Sci. USA 2014, 111, 14595–14600. [Google Scholar] [CrossRef] [PubMed]
- Seo, P.J.; Park, M.-J.; Lim, M.-H.; Kim, S.-G.; Lee, M.; Baldwin, I.T.; Park, C.-M. A self-regulatory circuit of circadian clock-associated1 underlies the circadian clock regulation of temperature responses in Arabidopsis. Plant Cell 2012, 24, 2427–2442. [Google Scholar] [CrossRef] [PubMed]
- James, A.B.; Syed, N.H.; Bordage, S.; Marshall, J.; Nimmo, G.A.; Jenkins, G.I.; Herzyk, P.; Brown, J.W.S.; Nimmo, H.G. Alternative splicing mediates responses of the Arabidopsis circadian clock to temperature changes. Plant Cell 2012, 24, 961–981. [Google Scholar] [CrossRef] [PubMed]
- Marshall, C.M.; Tartaglio, V.; Duarte, M.; Harmon, F.G. The Arabidopsis sickle mutant exhibits altered circadian clock responses to cool temperatures and temperature-dependent alternative splicing. Plant Cell 2016, 28, 2560–2575. [Google Scholar] [CrossRef] [PubMed]
- Johansson, M.; Staiger, D. Time to flower: Interplay between photoperiod and the circadian clock. J. Exp. Bot. 2015, 66, 719–730. [Google Scholar] [CrossRef] [PubMed]
- Shim, J.S.; Kubota, A.; Imaizumi, T. Circadian clock and photoperiodic flowering in Arabidopsis: CONSTANS is a hub for signal integration. Plant Physiol. 2017, 173, 5–15. [Google Scholar] [CrossRef] [PubMed]
- Nakamichi, N.; Kiba, T.; Kamioka, M.; Suzuki, T.; Yamashino, T.; Higashiyama, T.; Sakakibara, H.; Mizuno, T. Transcriptional repressor PRR5 directly regulates clock-output pathways. Proc. Natl. Acad. Sci. USA 2012, 109, 17123–17128. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Niwa, Y.; Ito, S.; Nakamichi, N.; Mizoguchi, T.; Niinuma, K.; Yamashino, T.; Mizuno, T. Genetic linkages of the circadian clock-associated genes, TOC1, CCA1 and LHY, in the photoperiodic control of flowering time in Arabidopsis thaliana. Plant Cell Physiol. 2007, 48, 925–937. [Google Scholar] [CrossRef] [PubMed]
- Song, Y.H.; Smith, R.W.; To, B.J.; Millar, A.J.; Imaizumi, T. FKF1 conveys timing information for CONSTANS stabilization in photoperiodic flowering. Science 2012, 336, 1045–1049. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hayama, R.; Sarid-Krebs, L.; Richter, R.; Fernández, V.; Jang, S.; Coupland, G. Pseudo response regulators stabilize constans protein to promote flowering in response to day length. EMBO J. 2017, 36, 904–918. [Google Scholar] [CrossRef] [PubMed]
- Nusinow, D.A.; Helfer, A.; Hamilton, E.E.; King, J.J.; Imaizumi, T.; Schultz, T.F.; Farre, E.M.; Kay, S.A. The ELF4-ELF3-LUX complex links the circadian clock to diurnal control of hypocotyl growth. Nature 2011, 475, 398–402. [Google Scholar] [CrossRef] [PubMed]
- Ezer, D.; Jung, J.-H.; Lan, H.; Biswas, S.; Gregoire, L.; Box, M.S.; Charoensawan, V.; Cortijo, S.; Lai, X.; Stöckle, D.; et al. The evening complex coordinates environmental and endogenous signals in Arabidopsis. Nat. Plants 2017, 3, 17087. [Google Scholar] [CrossRef] [PubMed]
- Nieto, C.; López-Salmerón, V.; Davière, J.-M.; Prat, S. ELF3-PIF4 interaction regulates plant growth independently of the evening complex. Curr. Boil. 2015, 25, 187–193. [Google Scholar] [CrossRef] [PubMed]
- Yoshida, R.; Fekih, R.; Fujiwara, S.; Oda, A.; Miyata, K.; Tomozoe, Y.; Nakagawa, M.; Niinuma, K.; Hayashi, K.; Ezura, H.; et al. Possible role of early flowering 3 (ELF3) in clock-dependent floral regulation by short vegetative phase (SVP) in Arabidopsis thaliana. New Phytol. 2009, 182, 838–850. [Google Scholar] [CrossRef] [PubMed]
- Yeom, M.; Kim, H.; Lim, J.; Shin, A.-Y.; Hong, S.; Kim, J.-I.; Nam, H.G. How do phytochromes transmit the light quality information to the circadian clock in Arabidopsis ? Mol. Plant 2014, 7, 1701–1704. [Google Scholar] [CrossRef] [PubMed]
- Huang, H.; Alvarez, S.; Bindbeutel, R.; Shen, Z.; Naldrett, M.J.; Evans, B.S.; Briggs, S.P.; Hicks, L.M.; Kay, S.A.; Nusinow, D.A. Identification of evening complex associated proteins in Arabidopsis by affinity purification and mass spectrometry. Mol. Cell. Proteom. 2016, 15, 201–217. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.L.; Covington, M.F.; Fankhauser, C.; Chory, J.; Wagner, D.R. ELF3 encodes a circadian clock-regulated nuclear protein that functions in an Arabidopsis phyb signal transduction pathway. Plant Cell 2001, 13, 1293–1304. [Google Scholar] [CrossRef] [PubMed]
- Reed, J.W.; Nagpal, P.; Bastow, R.M.; Solomon, K.S.; Dowson-Day, M.J.; Elumalai, R.P.; Millar, A.J. Independent action of ELF3 and phyb to control hypocotyl elongation and flowering time. Plant Physiol. 2000, 122, 1149–1160. [Google Scholar] [CrossRef] [PubMed]
- Wolters, H.; Jürgens, G. Survival of the flexible: Hormonal growth control and adaptation in plant development. Nat. Rev. Genet. 2009, 10, 305. [Google Scholar] [CrossRef] [PubMed]
- Davis, S.J. Integrating hormones into the floral-transition pathway of Arabidopsis thaliana. Plant Cell Environ. 2009, 32, 1201–1210. [Google Scholar] [CrossRef] [PubMed]
- Kazan, K.; Lyons, R. The link between flowering time and stress tolerance. J. Exp. Bot. 2015, 67, 47–60. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Galvão, V.C.; Schmid, M. Regulation of flowering by endogenous signals. In Advances in Botanical Research; Elsevier: Amsterdam, The Netherlands, 2014; Volume 72, pp. 63–102. [Google Scholar]
- Wilson, R.N.; Heckman, J.W.; Somerville, C.R. Gibberellin is required for flowering in Arabidopsis thaliana under short days. Plant Physiol. 1992, 100, 403–408. [Google Scholar] [CrossRef] [PubMed]
- Harberd, N.P. Relieving della restraint. Science 2003, 299, 1853–1854. [Google Scholar] [CrossRef] [PubMed]
- Davière, J.-M.; Achard, P. Gibberellin signaling in plants. Development 2013, 140, 1147–1151. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Conti, L. Hormonal control of the floral transition: Can one catch them all? Dev. Boil. 2017, 430, 288–301. [Google Scholar] [CrossRef] [PubMed]
- Davière, J.-M.; Achard, P. A pivotal role of dellas in regulating multiple hormone signals. Mol. Plant 2016, 9, 10–20. [Google Scholar] [CrossRef] [PubMed]
- de Lucas, M.; Davière, J.-M.; Rodríguez-Falcón, M.; Pontin, M.; Iglesias-Pedraz, J.M.; Lorrain, S.; Fankhauser, C.; Blázquez, M.A.; Titarenko, E.; Prat, S. A molecular framework for light and gibberellin control of cell elongation. Nature 2008, 451, 480–484. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Feng, S.; Martinez, C.; Gusmaroli, G.; Wang, Y.; Zhou, J.; Wang, F.; Chen, L.; Yu, L.; Iglesias-Pedraz, J.M.; Kircher, S.; et al. Coordinated regulation of Arabidopsis thaliana development by light and gibberellins. Nature 2008, 451, 475–479. [Google Scholar] [CrossRef] [PubMed]
- Galvão, V.C.; Collani, S.; Horrer, D.; Schmid, M. Gibberellic acid signaling is required for ambient temperature-mediated induction of flowering in Arabidopsis thaliana. Plant J. 2015, 84, 949–962. [Google Scholar] [CrossRef] [PubMed]
- Li, K.; Yu, R.; Fan, L.-M.; Wei, N.; Chen, H.; Deng, X.W. Della-mediated pif degradation contributes to coordination of light and gibberellin signalling in Arabidopsis. Nat. Commun. 2016, 7, 11868. [Google Scholar] [CrossRef] [PubMed]
- Yu, D.; Hu, Y.; Wang, H.; Pan, J.; Li, Y.; Lou, D. The della-constans transcription factor cascade integrates gibberelic acid and photoperiod signaling to regulate flowering. Plant Physiol. 2016, 172, 479–488. [Google Scholar]
- Chang, M.-Z.; Huang, C.-H. Effects of GA3 on promotion of flowering in Kalanchoe spp. Sci. Hortic. 2018, 238, 7–13. [Google Scholar] [CrossRef]
- Nolan, T.; Chen, J.; Yin, Y. Cross-talk of brassinosteroid signaling in controlling growth and stress responses. Biochem. J. 2017, 474, 2641–2661. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.-Y.; Bai, M.-Y.; Oh, E.; Zhu, J.-Y. Brassinosteroid signaling network and regulation of photomorphogenesis. Annu. Rev. Genet. 2012, 46, 701–724. [Google Scholar] [CrossRef] [PubMed]
- Ibañez, C.; Delker, C.; Martinez, C.; Bürstenbinder, K.; Janitza, P.; Lippmann, R.; Ludwig, W.; Sun, H.; James, G.V.; Klecker, M.; et al. Brassinosteroids dominate hormonal regulation of plant thermomorphogenesis via BZR1. Curr. Boil. 2018, 28, 303–310. [Google Scholar] [CrossRef] [PubMed]
- Martins, S.; Montiel-Jorda, A.; Cayrel, A.; Huguet, S.; Roux, C.P.-L.; Ljung, K.; Vert, G. Brassinosteroid signaling-dependent root responses to prolonged elevated ambient temperature. Nat. Commun. 2017, 8, 309. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jiang, W.B.; Huang, H.Y.; Hu, Y.W.; Zhu, S.W.; Wang, Z.Y.; Lin, W.H. Brassinosteroid regulates seed size and shape in Arabidopsis. Plant Physiol. 2013, 162, 1965–1977. [Google Scholar] [CrossRef] [PubMed]
- Oh, E.; Zhu, J.-Y.; Wang, Z.-Y. Interaction between BZR1 and PIF4 integrates brassinosteroid and environmental responses. Nat. Cell Boil. 2012, 14, 802–809. [Google Scholar] [CrossRef] [PubMed]
- Belkhadir, Y.; Jaillais, Y. The molecular circuitry of brassinosteroid signaling. New Phytol. 2015, 206, 522–540. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, Z.; Ou, Y.; Zhang, Z.; Li, J.; He, Y. Brassinosteroid signaling recruits histone 3 lysine-27 demethylation activity to Flowering Locus C chromatin to inhibit the floral transition in Arabidopsis. Mol. Plant 2018, 11, 1135–1146. [Google Scholar] [CrossRef] [PubMed]
- Domagalska, M.A.; Schomburg, F.M.; Amasino, R.M.; Vierstra, R.D.; Nagy, F.; Davis, S.J. Attenuation of brassinosteroid signaling enhances FLC expression and delays flowering. Development 2007, 134, 2841–2850. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Y.; Li, B.; Xu, Y.; Li, H.; Li, S.; Zhang, D.; Mao, Z.; Guo, S.; Yang, C.; Weng, Y.; et al. The cyclophilin CYP20-2 modulates the conformation of BRASSINAZOLE-RESISTANT1, which binds the promoter of FLOWERING LOCUS D to regulate flowering in Arabidopsis. Plant Cell 2013, 25, 2504–2521. [Google Scholar] [CrossRef] [PubMed]
- Crevillén, P.; Yang, H.; Cui, X.; Greeff, C.; Trick, M.; Qiu, Q.; Cao, X.; Dean, C. Epigenetic reprogramming that prevents transgenerational inheritance of the vernalized state. Nature 2014, 515, 587–590. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Browse, J. Jasmonate passes muster: A receptor and targets for the defense hormone. Annu. Rev. Plant Boil. 2009, 60, 183–205. [Google Scholar] [CrossRef] [PubMed]
- Krajnčič, B.; Kristl, J.; Janžekovič, I. Possible role of jasmonic acid in the regulation of floral induction, evocation and floral differentiation in Lemna minor L. Plant Physiol. Biochem. 2006, 44, 752–758. [Google Scholar] [CrossRef] [PubMed]
- Robson, F.; Okamoto, H.; Patrick, E.; Harris, S.-R.; Wasternack, C.; Brearley, C.; Turner, J.G. Jasmonate and phytochrome A signaling in Arabidopsis wound and shade responses are integrated through JAZ1 stability. Plant Cell 2010, 22, 1143–1160. [Google Scholar] [CrossRef] [PubMed]
- Chini, A.; Fonseca, S.; Fernandez, G.; Adie, B.; Chico, J.; Lorenzo, O.; Garcia-Casado, G.; Lopez-Vidriero, I.; Lozano, F.; Ponce, M. The JAZ family of repressors is the missing link in jasmonate signalling. Nature 2007, 448, 666–671. [Google Scholar] [CrossRef] [PubMed]
- Thines, B.; Katsir, L.; Melotto, M.; Niu, Y.; Mandaokar, A.; Liu, G.; Nomura, K.; He, S.Y.; Howe, G.A.; Browse, J. JAZ repressor proteins are targets of the SCFCOI1 complex during jasmonate signalling. Nature 2007, 448, 661–665. [Google Scholar] [CrossRef] [PubMed]
- Zhai, Q.; Zhang, X.; Wu, F.; Feng, H.; Deng, L.; Xu, L.; Zhang, M.; Wang, Q.; Li, C. Transcriptional mechanism of jasmonate receptor COI1-mediated delay of flowering time in Arabidopsis. Plant Cell 2015. [Google Scholar] [CrossRef] [PubMed]
- Hibara, K.-I.; Isono, M.; Mimura, M.; Sentoku, N.; Kojima, M.; Sakakibara, H.; Kitomi, Y.; Yoshikawa, T.; Itoh, J.-I.; Nagato, Y. Jasmonate regulates juvenile-adult phase transition in rice. Development 2016, 143, 3407–3416. [Google Scholar] [CrossRef] [PubMed]




© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Susila, H.; Nasim, Z.; Ahn, J.H. Ambient Temperature-Responsive Mechanisms Coordinate Regulation of Flowering Time. Int. J. Mol. Sci. 2018, 19, 3196. https://doi.org/10.3390/ijms19103196
Susila H, Nasim Z, Ahn JH. Ambient Temperature-Responsive Mechanisms Coordinate Regulation of Flowering Time. International Journal of Molecular Sciences. 2018; 19(10):3196. https://doi.org/10.3390/ijms19103196
Chicago/Turabian StyleSusila, Hendry, Zeeshan Nasim, and Ji Hoon Ahn. 2018. "Ambient Temperature-Responsive Mechanisms Coordinate Regulation of Flowering Time" International Journal of Molecular Sciences 19, no. 10: 3196. https://doi.org/10.3390/ijms19103196
APA StyleSusila, H., Nasim, Z., & Ahn, J. H. (2018). Ambient Temperature-Responsive Mechanisms Coordinate Regulation of Flowering Time. International Journal of Molecular Sciences, 19(10), 3196. https://doi.org/10.3390/ijms19103196