Nonlinear Cellular Mechanical Behavior Adaptation to Substrate Mechanics Identified by Atomic Force Microscope
Abstract
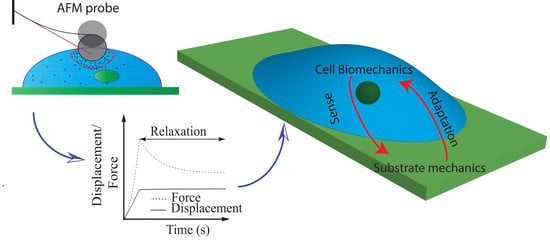
:1. Introduction
2. Results and Discussion
2.1. Effect of Substrate Mechanics on the Nonlinear Elastic and Viscoelastic Behavior of the Cells
2.2. Effect of Substrate Mechanics on the Poroelastic Behavior of the Cells
2.3. Substrate Mechanics Affects Cell Biomechanical Behavior by Regulating the Cell Morphology
3. Conclusions
4. Materials and Methods
4.1. Chemicals
4.2. Polymer Substrate Preparation
4.3. Cell Culture and Treatment
4.4. Immunofluorescence and F-Actin Quantification
4.5. Atomic Force Microscopy (AFM) Measurement
4.6. Biomechanical Quantification of the Cells
4.6.1. Cell Elasticity
4.6.2. Viscoelasticity
4.6.3. Poroelasticity
4.7. Stiffness Quantification of the Substrates
4.8. Curve Fitting and Statistical Analysis
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Leipzig, N.D.; Shoichet, M.S. The effect of substrate stiffness on adult neural stem cell behavior. Biomaterials 2009, 30, 6867–6878. [Google Scholar] [CrossRef] [PubMed]
- Yeung, T.; Georges, P.C.; Flanagan, L.A.; Marg, B.; Ortiz, M.; Funaki, M.; Zahir, N.; Ming, W.; Weaver, V.; Janmey, P.A. Effects of substrate stiffness on cell morphology, cytoskeletal structure, and adhesion. Cytoskeleton 2005, 60, 24–34. [Google Scholar] [CrossRef] [PubMed]
- Brandl, F.; Sommer, F.; Goepferich, A. Rational design of hydrogels for tissue engineering: Impact of physical factors on cell behavior. Biomaterials 2007, 28, 134–146. [Google Scholar] [CrossRef] [PubMed]
- Kilian, K.A.; Bugarija, B.; Lahn, B.T.; Mrksich, M. Geometric cues for directing the differentiation of mesenchymal stem cells. Proc. Natl. Acad. Sci. USA 2010, 107, 4872–4877. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guilak, F.; Cohen, D.M.; Estes, B.T.; Gimble, J.M.; Liedtke, W.; Chen, C.S. Control of stem cell fate by physical interactions with the extracellular matrix. Cell Stem Cell 2009, 5, 17–26. [Google Scholar] [CrossRef] [PubMed]
- Ulrich, T.A.; De Juan Pardo, E.M.; Kumar, S. The mechanical rigidity of the extracellular matrix regulates the structure, motility, and proliferation of glioma cells. Cancer Res. 2009, 69, 4167–4174. [Google Scholar] [CrossRef] [PubMed]
- Jiang, G.; Huang, A.H.; Cai, Y.; Tanase, M.; Sheetz, M.P. Rigidity sensing at the leading edge through αvβ3 integrins and RPTPα. Biophys. J. 2006, 90, 1804–1809. [Google Scholar] [CrossRef] [PubMed]
- Giannone, G.; Sheetz, M.P. Substrate rigidity and force define form through tyrosine phosphatase and kinase pathways. Trends Cell Biol. 2006, 16, 213–223. [Google Scholar] [CrossRef] [PubMed]
- Efremov, Y.M.; Wang, W.H.; Hardy, S.D.; Geahlen, R.L.; Raman, A. Measuring nanoscale viscoelastic parameters of cells directly from AFM force-displacement curves. Sci. Rep. 2017, 7, 1541. [Google Scholar] [CrossRef] [PubMed]
- Xie, S.; Ren, J. Note: Precision control of nano-positioning stage: An iterative learning-based model predictive control approach. Rev. Sci. Instrum. 2018, 89, 076103. [Google Scholar] [CrossRef] [PubMed]
- Discher, D.E.; Janmey, P.; Wang, Y.L. Tissue cells feel and respond to the stiffness of their substrate. Science 2005, 310, 1139–1143. [Google Scholar] [CrossRef] [PubMed]
- Rianna, C.; Radmacher, M. Influence of microenvironment topography and stiffness on the mechanics and motility of normal and cancer renal cells. Nanoscale 2017, 9, 11222–11230. [Google Scholar] [CrossRef] [PubMed]
- Tee, S.Y.; Fu, J.; Chen, C.S.; Janmey, P.A. Cell shape and substrate rigidity both regulate cell stiffness. Biophys. J. 2011, 100, L25–L27. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.B.; Dembo, M.; Wang, Y.L. Substrate flexibility regulates growth and apoptosis of normal but not transformed cells. Am. J. Physiol.-Cell Physiol. 2000, 279, C1345–C1350. [Google Scholar] [CrossRef] [PubMed]
- Takai, E.; Costa, K.D.; Shaheen, A.; Hung, C.T.; Guo, X.E. Osteoblast elastic modulus measured by atomic force microscopy is substrate dependent. Ann. Biomed. Eng. 2005, 33, 963–971. [Google Scholar] [CrossRef] [PubMed]
- Mollaeian, K.; Liu, Y.; Bi, S.; Ren, J. Atomic force microscopy study revealed velocity-dependence and nonlinearity of nanoscale poroelasticity of eukaryotic cells. J. Mech. Behav. Biomed. Mater. 2018, 78, 65–73. [Google Scholar] [CrossRef] [PubMed]
- Moeendarbary, E.; Valon, L.; Fritzsche, M.; Harris, A.R.; Moulding, D.A.; Thrasher, A.J.; Stride, E.; Mahadevan, L.; Charras, G.T. The cytoplasm of living cells behaves as a poroelastic material. Nat. Mater. 2013, 12, 253–261. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nia, H.T.; Han, L.; Li, Y.; Ortiz, C.; Grodzinsky, A. Poroelasticity of cartilage at the nanoscale. Biophys. J. 2011, 101, 2304–2313. [Google Scholar] [CrossRef] [PubMed]
- Charras, G.T.; Yarrow, J.C.; Horton, M.A.; Mahadevan, L.; Mitchison, T. Non-equilibration of hydrostatic pressure in blebbing cells. Nature 2005, 435, 365–369. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Charras, G.T.; Mitchison, T.J.; Mahadevan, L. Animal cell hydraulics. J. Cell Sci. 2009, 122, 3233–3241. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dembo, M.; Harlow, F. Cell motion, contractile networks, and the physics of interpenetrating reactive flow. Biophys. J. 1986, 50, 109–121. [Google Scholar] [CrossRef] [Green Version]
- Charras, G.T.; Coughlin, M.; Mitchison, T.J.; Mahadevan, L. Life and times of a cellular bleb. Biophys. J. 2008, 94, 1836–1853. [Google Scholar] [CrossRef] [PubMed]
- Solon, J.; Levental, I.; Sengupta, K.; Georges, P.C.; Janmey, P.A. Fibroblast adaptation and stiffness matching to soft elastic substrates. Biophys. J. 2007, 93, 4453–4461. [Google Scholar] [CrossRef] [PubMed]
- Sneddon, I.N. The relation between load and penetration in the axisymmetric Boussinesq problem for a punch of arbitrary profile. Int. J. Eng. Sci. 1965, 3, 47–57. [Google Scholar] [CrossRef]
- Ghaednia, H.; Wang, X.; Saha, S.; Xu, Y.; Sharma, A.; Jackson, R.L. A review of elastic–plastic contact mechanics. Appl. Mech. Rev. 2017, 69, 060804. [Google Scholar] [CrossRef]
- Wang, H.B.; Dembo, M.; Hanks, S.K.; Wang, Y.L. Focal adhesion kinase is involved in mechanosensing during fibroblast migration. Proc. Natl. Acad. Sci. USA 2001, 98, 11295–11300. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ghibaudo, M.; Saez, A.; Trichet, L.; Xayaphoummine, A.; Browaeys, J.; Silberzan, P.; Buguin, A.; Ladoux, B. Traction forces and rigidity sensing regulate cell functions. Soft Matter 2008, 4, 1836–1843. [Google Scholar] [CrossRef]
- Saez, A.; Buguin, A.; Silberzan, P.; Ladoux, B. Is the mechanical activity of epithelial cells controlled by deformations or forces? Biophys. J. 2005, 89, L52–L54. [Google Scholar] [CrossRef] [PubMed]
- Deuschle, J.K.; De Souza, E.J.; Arzt, E.; Enders, S. Nanoindentation studies on crosslinking and curing effects of PDMS. Int. J. Mater. Res. 2010, 101, 1014–1023. [Google Scholar] [CrossRef]
- Carrillo, F.; Gupta, S.; Balooch, M.; Marshall, S.J.; Marshall, G.W.; Pruitt, L.; Puttlitz, C.M. Nanoindentation of polydimethylsiloxane elastomers: Effect of crosslinking, work of adhesion, and fluid environment on elastic modulus. J. Mater. Res. 2005, 20, 2820–2830. [Google Scholar] [CrossRef]
- Alisafaei, F.; Han, C.S.; Sanei, S.H.R. On the time and indentation depth dependence of hardness, dissipation and stiffness in polydimethylsiloxane. Polym. Test. 2013, 32, 1220–1228. [Google Scholar] [CrossRef]
- Charitidis, C.A. Nanoscale deformation and nanomechanical properties of polydimethylsiloxane (PDMS). Ind. Eng. Chem. Res. 2010, 50, 565–570. [Google Scholar] [CrossRef]
- Wang, Z.; Volinsky, A.A.; Gallant, N.D. Crosslinking effect on polydimethylsiloxane elastic modulus measured by custom-built compression instrument. J. Appl. Polym. Sci. 2014, 131. [Google Scholar] [CrossRef] [Green Version]
- Pogoda, K.; Jaczewska, J.; Wiltowska-Zuber, J.; Klymenko, O.; Zuber, K.; Fornal, M.; Lekka, M. Depth-sensing analysis of cytoskeleton organization based on AFM data. Eur. Biophys. J. 2012, 41, 79–87. [Google Scholar] [CrossRef] [PubMed]
- Schillers, H.; Wälte, M.; Urbanova, K.; Oberleithner, H. Real-time monitoring of cell elasticity reveals oscillating myosin activity. Biophys. J. 2010, 99, 3639–3646. [Google Scholar] [CrossRef] [PubMed]
- Sirghi, L.; Ponti, J.; Broggi, F.; Rossi, F. Probing elasticity and adhesion of live cells by atomic force microscopy indentation. Eur. Biophys. J. 2008, 37, 935–945. [Google Scholar] [CrossRef] [PubMed]
- Park, J.S.; Chu, J.S.; Tsou, A.D.; Diop, R.; Tang, Z.; Wang, A.; Li, S. The effect of matrix stiffness on the differentiation of mesenchymal stem cells in response to TGF-β. Biomaterials 2011, 32, 3921–3930. [Google Scholar] [CrossRef] [PubMed]
- Guo, W.H.; Frey, M.T.; Burnham, N.A.; Wang, Y.L. Substrate rigidity regulates the formation and maintenance of tissues. Biophys. J. 2006, 90, 2213–2220. [Google Scholar] [CrossRef] [PubMed]
- Engler, A.J.; Griffin, M.A.; Sen, S.; Bönnemann, C.G.; Sweeney, H.L.; Discher, D.E. Myotubes differentiate optimally on substrates with tissue-like stiffness: Pathological implications for soft or stiff microenvironments. J. Cell Biol. 2004, 166, 877–887. [Google Scholar] [CrossRef] [PubMed]
- McNaught, A.D.; Wilkinson, A. Compendium of Chemical Terminology; Blackwell Science: Oxford, UK, 1997; Volume 1669. [Google Scholar]
- Burridge, K.; Fath, K.; Kelly, T.; Nuckolls, G.; Turner, C. Focal adhesions: transmembrane junctions between the extracellular matrix and the cytoskeleton. Ann. Rev. Cell Biol. 1988, 4, 487–525. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.S.; Tan, J.; Tien, J. Mechanotransduction at cell-matrix and cell-cell contacts. Annu. Rev. Biomed. Eng. 2004, 6, 275–302. [Google Scholar] [CrossRef] [PubMed]
- Geiger, B.; Spatz, J.P.; Bershadsky, A.D. Environmental sensing through focal adhesions. Nat. Rev. Mol. Cell Biol. 2009, 10, 21. [Google Scholar] [CrossRef] [PubMed]
- Yim, E.K.; Darling, E.M.; Kulangara, K.; Guilak, F.; Leong, K.W. Nanotopography-induced changes in focal adhesions, cytoskeletal organization, and mechanical properties of human mesenchymal stem cells. Biomaterials 2010, 31, 1299–1306. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Munson, B.R.; Okiishi, T.H.; Rothmayer, A.P.; Huebsch, W.W. Fundamentals of Fluid Mechanics; John Wiley & Sons: Hoboken, NJ, USA, 2014. [Google Scholar]
- Wells, R.E.; Merrill, E.W. Shear rate dependence of the viscosity of whole blood and plasma. Science 1961, 133, 763–764. [Google Scholar] [CrossRef] [PubMed]
- Sorce, B.; Escobedo, C.; Toyoda, Y.; Stewart, M.P.; Cattin, C.J.; Newton, R.; Banerjee, I.; Stettler, A.; Roska, B.; Eaton, S.; et al. Mitotic cells contract actomyosin cortex and generate pressure to round against or escape epithelial confinement. Nat. Commun. 2015, 6, 8872. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gupta, M.; Sarangi, B.R.; Deschamps, J.; Nematbakhsh, Y.; Callan-Jones, A.; Margadant, F.; Mège, R.M.; Lim, C.T.; Voituriez, R.; Ladoux, B. Adaptive rheology and ordering of cell cytoskeleton govern matrix rigidity sensing. Nat. Commun. 2015, 6, 7525. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jin, M.; Andricioaei, I.; Springer, T.A. Conversion between three conformational states of integrin I domains with a C-terminal pull spring studied with molecular dynamics. Structure 2004, 12, 2137–2147. [Google Scholar] [CrossRef] [PubMed]
- Hinz, B.; Celetta, G.; Tomasek, J.J.; Gabbiani, G.; Chaponnier, C. Alpha-smooth muscle actin expression upregulates fibroblast contractile activity. Mol. Biol. Cell 2001, 12, 2730–2741. [Google Scholar] [CrossRef] [PubMed]
- Chrzanowska-Wodnicka, M.; Burridge, K. Rho-stimulated contractility drives the formation of stress fibers and focal adhesions. J. Cell Biol. 1996, 133, 1403–1415. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Beningo, K.A.; Wang, Y.L. Fc-receptor-mediated phagocytosis is regulated by mechanical properties of the target. J. Cell Sci. 2002, 115, 849–856. [Google Scholar] [PubMed]
- Pelham, R.J.; Wang, Y.l. Cell locomotion and focal adhesions are regulated by substrate flexibility. Proc. Natl. Acad. Sci. USA 1997, 94, 13661–13665. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Doyle, A.; Marganski, W.; Lee, J. Calcium transients induce spatially coordinated increases in traction force during the movement of fish keratocytes. J. Cell Sci. 2004, 117, 2203–2214. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fillingham, I.; Gingras, A.R.; Papagrigoriou, E.; Patel, B.; Emsley, J.; Critchley, D.R.; Roberts, G.C.; Barsukov, I.L. A vinculin binding domain from the talin rod unfolds to form a complex with the vinculin head. Structure 2005, 13, 65–74. [Google Scholar] [CrossRef] [PubMed]
- Alapan, Y.; Younesi, M.; Akkus, O.; Gurkan, U.A. Anisotropically stiff 3D micropillar niche induces extraordinary cell alignment and elongation. Adv. Healthc. Mater. 2016, 5, 1884–1892. [Google Scholar] [CrossRef] [PubMed]
- Marelli, M.; Gadhari, N.; Boero, G.; Chiquet, M.; Brugger, J. Cell force measurements in 3D microfabricated environments based on compliant cantilevers. Lab Chip 2014, 14, 286–293. [Google Scholar] [CrossRef] [PubMed]
- Sniadecki, N.J.; Anguelouch, A.; Yang, M.T.; Lamb, C.M.; Liu, Z.; Kirschner, S.B.; Liu, Y.; Reich, D.H.; Chen, C.S. Magnetic microposts as an approach to apply forces to living cells. Proc. Natl. Acad. Sci. USA 2007, 104, 14553–14558. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hutter, J.L.; Bechhoefer, J. Calibration of atomic-force microscope tips. Rev. Sci. Instrum. 1993, 64, 1868–1873. [Google Scholar] [CrossRef]
- Dimitriadis, E.K.; Horkay, F.; Maresca, J.; Kachar, B.; Chadwick, R.S. Determination of elastic moduli of thin layers of soft material using the atomic force microscope. Biophys. J. 2002, 82, 2798–2810. [Google Scholar] [CrossRef]
- Charras, G.; Lehenkari, P.P.; Horton, M. Atomic force microscopy can be used to mechanically stimulate osteoblasts and evaluate cellular strain distributions. Ultramicroscopy 2001, 86, 85–95. [Google Scholar] [CrossRef]
- Chan, E.P.; Hu, Y.; Johnson, P.M.; Suo, Z.; Stafford, C.M. Spherical indentation testing of poroelastic relaxations in thin hydrogel layers. Soft Matter 2012, 8, 1492–1498. [Google Scholar] [CrossRef]
- Mollaeian, K.; Liu, Y.; Ren, J. Investigation of nanoscale poroelasticity of eukaryotic cells using Atomic Force Microscopy. In Proceedings of the ASME 2017 Dynamic Systems and Control Conference, Tysons Corner, VA, USA, 11–13 October 2017; p. V001T08A005. [Google Scholar]
- Ren, J.; Huang, H.; Liu, Y.; Zheng, X.; Zou, Q. An atomic force microscope study revealed two mechanisms in the effect of anticancer drugs on rate-dependent Young’a modulus of human prostate cancer cells. PLoS ONE 2015, 10, e0126107. [Google Scholar]
- Kalcioglu, Z.I.; Mahmoodian, R.; Hu, Y.; Suo, Z.; Van Vliet, K.J. From macro-to microscale poroelastic characterization of polymeric hydrogels via indentation. Soft Matter 2012, 8, 3393–3398. [Google Scholar] [CrossRef]
- Ghaednia, H.; Pope, S.A.; Jackson, R.L.; Marghitu, D.B. A comprehensive study of the elasto-plastic contact of a sphere and a flat. Tribol. Int. 2016, 93, 78–90. [Google Scholar] [CrossRef]
- Liu, Y.; Mollaeian, K.; Ren, J. Finite element modeling of living cells for AFM indentation-based biomechanical characterization. Micron 2019, 116, 108–115. [Google Scholar] [CrossRef] [PubMed]
- Darling, E.M.; Zauscher, S.; Block, J.A.; Guilak, F. A thin-layer model for viscoelastic, stress-relaxation testing of cells using atomic force microscopy: Do cell properties reflect metastatic potential? Biophys. J. 2007, 92, 1784–1791. [Google Scholar] [CrossRef] [PubMed]
- Ketene, A.N.; Schmelz, E.M.; Roberts, P.C.; Agah, M. The effects of cancer progression on the viscoelasticity of ovarian cell cytoskeleton structures. Nanomed. Nanotechnol. Biol. Med. 2012, 8, 93–102. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.; Zhao, X.; Vlassak, J.J.; Suo, Z. Using indentation to characterize the poroelasticity of gels. Appl. Phys. Lett. 2010, 96, 121904. [Google Scholar] [CrossRef]





© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mollaeian, K.; Liu, Y.; Bi, S.; Wang, Y.; Ren, J.; Lu, M. Nonlinear Cellular Mechanical Behavior Adaptation to Substrate Mechanics Identified by Atomic Force Microscope. Int. J. Mol. Sci. 2018, 19, 3461. https://doi.org/10.3390/ijms19113461
Mollaeian K, Liu Y, Bi S, Wang Y, Ren J, Lu M. Nonlinear Cellular Mechanical Behavior Adaptation to Substrate Mechanics Identified by Atomic Force Microscope. International Journal of Molecular Sciences. 2018; 19(11):3461. https://doi.org/10.3390/ijms19113461
Chicago/Turabian StyleMollaeian, Keyvan, Yi Liu, Siyu Bi, Yifei Wang, Juan Ren, and Meng Lu. 2018. "Nonlinear Cellular Mechanical Behavior Adaptation to Substrate Mechanics Identified by Atomic Force Microscope" International Journal of Molecular Sciences 19, no. 11: 3461. https://doi.org/10.3390/ijms19113461
APA StyleMollaeian, K., Liu, Y., Bi, S., Wang, Y., Ren, J., & Lu, M. (2018). Nonlinear Cellular Mechanical Behavior Adaptation to Substrate Mechanics Identified by Atomic Force Microscope. International Journal of Molecular Sciences, 19(11), 3461. https://doi.org/10.3390/ijms19113461