Vascular Plant One-Zinc-Finger (VOZ) Transcription Factors Are Positive Regulators of Salt Tolerance in Arabidopsis
Abstract
:1. Introduction
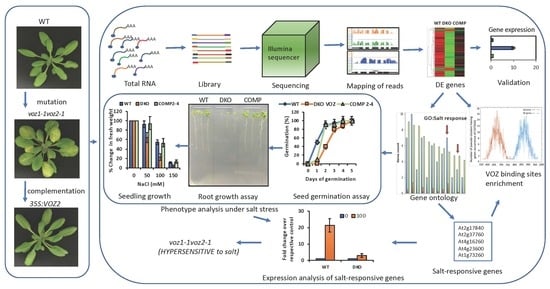
2. Results
2.1. Loss of Function of VOZs Resulted in Misregulation of Genes
2.2. VOZs Regulate Expression of Several Transcription Factors
2.3. Promoters of Differentially Expressed Genes are Enriched for G-Box, NAC and LS-7 Elements
2.4. GO Term Enrichment Analysis for Biological Processes in Differentially Expressed Genes
2.5. VOZs Regulate Expression of Many Abiotic Stress-Responsive Genes
2.6. VOZs Regulate Salt Stress Tolerance
2.6.1. VOZ Double Mutant Exhibits Hypersensitivity to Salt Stress
2.6.2. VOZs Activate the Expression of Salt-Responsive Genes
3. Discussion
3.1. VOZs are Involved in Regulation of Many Stress-Responsive Genes
3.2. Genes with Binding Motifs for VOZs are Both Up- and Down-Regulated
3.3. VOZs Likely Regulate Expression of Some Genes Indirectly
3.4. VOZ Confers Salt Tolerance by Activating the Expression of Salt-Responsive Genes
4. Materials and Methods
4.1. Plant Materials and Growth Conditions
4.2. Salt Stress Treatment
4.3. RNA-Seq
4.4. Mapping of the Reads and Identification of Differentially Expressed (DE) Genes
4.5. Bioinformatics Analysis of DE Genes for VOZs Binding Motifs
4.6. GO Term Enrichment Analysis
4.7. Identification of TFs, Abiotic Stress and Hormone-Responsive Genes in DE List
4.8. Promoter Analysis for Enrichment of Cis-Elements
4.9. Validation of DE Genes Using RT-qPCR Analysis
4.10. RT-qPCR Analysis of Salt-Responsive Genes
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
Abbreviation
References
- Cheeseman, J.M. The evolution of halophytes, glycophytes and crops, and its implications for food security under saline conditions. New Phytol. 2015, 206, 557–570. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.K. Abiotic stress signaling and responses in plants. Cell 2016, 167, 313–324. [Google Scholar] [CrossRef] [PubMed]
- Reddy, A.S.; Ali, G.S.; Celesnik, H.; Day, I.S. Coping with stresses: Roles of calcium- and calcium/calmodulin-regulated gene expression. Plant Cell 2011, 23, 2010–2032. [Google Scholar] [CrossRef] [PubMed]
- Ohama, N.; Sato, H.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Transcriptional Regulatory Network of Plant Heat Stress Response. Trends Plant Sci. 2017, 22, 53–65. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.S.; An, C.; Park, S.; Gilmour, S.J.; Wang, L.; Renna, L.; Brandizzi, F.; Grumet, R.; Thomashow, M.F. CAMTA-Mediated Regulation of Salicylic Acid Immunity Pathway Genes in Arabidopsis Exposed to Low Temperature and Pathogen Infection. Plant Cell 2017, 29, 2465–2477. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Prasad, K.V.S.K.; Abdel-Hameed, A.A.E.; Xing, D.; Reddy, A.S.N. Global gene expression analysis using RNA-seq uncovered a new role for SR1/CAMTA3 transcription factor in salt stress. Sci. Rep. 2016, 6, 27021. [Google Scholar] [CrossRef] [PubMed]
- Khan, S.A.; Li, M.Z.; Wang, S.M.; Yin, H.J. Revisiting the Role of Plant Transcription Factors in the Battle against Abiotic Stress. Int. J. Mol. Sci. 2018, 19, 1634. [Google Scholar] [CrossRef] [PubMed]
- Mitsuda, N.; Hisabori, T.; Takeyasu, K.; Sato, M.H. VOZ; isolation and characterization of novel vascular plant transcription factors with a one-zinc finger from Arabidopsis thaliana. Plant Cell Physiol. 2004, 45, 845–854. [Google Scholar] [CrossRef] [PubMed]
- Yasui, Y.; Mukougawa, K.; Uemoto, M.; Yokofuji, A.; Suzuri, R.; Nishitani, A.; Kohchi, T. The phytochrome-interacting vascular plant one-zinc finger1 and VOZ2 redundantly regulate flowering in Arabidopsis. Plant Cell 2012, 24, 3248–3263. [Google Scholar] [CrossRef] [PubMed]
- Jensen, M.K.; Kjaersgaard, T.; Nielsen, M.M.; Galberg, P.; Petersen, K.; O’Shea, C.; Skriver, K. The Arabidopsis thaliana NAC transcription factor family: Structure-function relationships and determinants of ANAC019 stress signalling. Biochem. J. 2010, 426, 183–196. [Google Scholar] [CrossRef] [PubMed]
- Celesnik, H.; Ali, G.S.; Robison, F.M.; Reddy, A.S. Arabidopsis thaliana VOZ (Vascular plant One-Zinc finger) transcription factors are required for proper regulation of flowering time. Biol. Open 2013, 2, 424–431. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kumar, S.; Choudhary, P.; Gupta, M.; Nath, U. VASCULAR PLANT ONE-ZINC FINGER1 (VOZ1) and VOZ2 Interact with CONSTANS and Promote Photoperiodic Flowering Transition. Plant Physiol. 2018, 176, 2917–2930. [Google Scholar] [CrossRef] [PubMed]
- Nakai, Y.; Fujiwara, S.; Kubo, Y.; Sato, M.H. Overexpression of VOZ2 confers biotic stress tolerance but decreases abiotic stress resistance in Arabidopsis. Plant Signal. Behav. 2013, 8, e23358. [Google Scholar] [CrossRef] [PubMed]
- Nakai, Y.; Nakahira, Y.; Sumida, H.; Takebayashi, K.; Nagasawa, Y.; Yamasaki, K.; Akiyama, M.; Ohme-Takagi, M.; Fujiwara, S.; Shiina, T.; et al. Vascular plant one-zinc-finger protein 1/2 transcription factors regulate abiotic and biotic stress responses in Arabidopsis. Plant J. 2013, 73, 761–775. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Koguchi, M.; Yamasaki, K.; Hirano, T.; Sato, M.H. Vascular plant one-zinc-finger protein 2 is localized both to the nucleus and stress granules under heat stress in Arabidopsis. Plant Signal. Behav. 2017, 12, e1295907. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Song, C.; Lee, J.; Kim, T.; Hong, J.C.; Lim, C.O. VOZ1, a transcriptional repressor of DREB2C, mediates heat stress responses in Arabidopsis. Planta 2018, 247, 1439–1448. [Google Scholar] [CrossRef] [PubMed]
- Yasui, Y.; Kohchi, T. VASCULAR PLANT ONE-ZINC FINGER1 and VOZ2 repress the FLOWERING LOCUS C clade members to control flowering time in Arabidopsis. Biosci. Biotechnol. Biochem. 2014, 78, 1850–1855. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Oberschall, A.; Deák, M.; Török, K.; Sass, L.; Vass, I.; Kovács, I.; Fehér, A.; Dudits, D.; Horváth, G.V. A novel aldose/aldehyde reductase protects transgenic plants against lipid peroxidation under chemical and drought stresses. Plant J. 2000, 24, 437–446. [Google Scholar] [CrossRef] [PubMed]
- Simpson, P.J.; Tantitadapitak, C.; Reed, A.M.; Mather, O.C.; Bunce, C.M.; White, S.A.; Ride, J.P. Characterization of Two Novel Aldo-Keto Reductases from Arabidopsis: Expression Patterns, Broad Substrate Specificity, and an Open Active-Site Structure Suggest a Role in Toxicant Metabolism Following Stress. J. Mol. Biol. 2009, 392, 465–480. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Li, F.; Wang, J.L.; Ma, Y.; Chong, K.; Xu, Y.Y. Basic helix-loop-helix transcription factor from wild rice (OrbHLH2) improves tolerance to salt- and osmotic stress in Arabidopsis. J. Plant Physiol. 2009, 166, 1296–1306. [Google Scholar] [CrossRef] [PubMed]
- Safi, H.; Saibi, W.; Alaoui, M.M.; Hmyene, A.; Masmoudi, K.; Hanin, M.; Brini, F. A wheat lipid transfer protein (TdLTP4) promotes tolerance to abiotic and biotic stress in Arabidopsis thaliana. Plant Physiol. Biochem. 2015, 89, 64–75. [Google Scholar] [CrossRef] [PubMed]
- Kim, I.J.L.; Yun, B.W.; Jamil, M. GA Mediated OsZAT-12 Expression Improves Salt Resistance of Rice. Int. J. Agric. Biol. 2016, 18, 330–336. [Google Scholar]
- Le, C.T.; Brumbarova, T.; Ivanov, R.; Stoof, C.; Weber, E.; Mohrbacher, J.; Fink-Straube, C.; Bauer, P. ZINC FINGER OF ARABIDOPSIS THALIANA12 (ZAT12) Interacts with FER-LIKE IRON DEFICIENCY-INDUCED TRANSCRIPTION FACTOR (FIT) Linking Iron Deficiency and Oxidative Stress Responses. Plant Physiol. 2016, 170, 540–557. [Google Scholar] [CrossRef] [PubMed]
- Vogel, J.T.; Zarka, D.G.; Van Buskirk, H.A.; Fowler, S.G.; Thomashow, M.F. Roles of the CBF2 and ZAT12 transcription factors in configuring the low temperature transcriptome of Arabidopsis. Plant J. 2005, 41, 195–211. [Google Scholar] [CrossRef] [PubMed]
- Gong, Z.; Koiwa, H.; Cushman, M.A.; Ray, A.; Bufford, D.; Kore-eda, S.; Matsumoto, T.K.; Zhu, J.; Cushman, J.C.; Bressan, R.A.; et al. Genes that are uniquely stress regulated in salt overly sensitive (sos) mutants. Plant Physiol. 2001, 126, 363–375. [Google Scholar] [CrossRef] [PubMed]
- Shan, L.; Li, C.L.; Chen, F.; Zhao, S.Y.; Xia, G.M. A Bowman-Birk type protease inhibitor is involved in the tolerance to salt stress in wheat. Plant Cell Environ. 2008, 31, 1128–1137. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, R.; Wang, W.J.; Wang, W.G.; Li, F.S.; Wang, Q.W.; Xu, Y.; Wang, S.H. Overexpression of a cysteine proteinase inhibitor gene from Jatropha curcas confers enhanced tolerance to salinity stress. Electron. J. Biotechnol. 2015, 18, 368–375. [Google Scholar] [CrossRef]
- Srinivasan, T.; Kumar, K.R.; Kirti, P.B. Constitutive expression of a trypsin protease inhibitor confers multiple stress tolerance in transgenic tobacco. Plant Cell Physiol. 2009, 50, 541–553. [Google Scholar] [CrossRef] [PubMed]
- Reddy, A.S.; Reddy, V.S.; Golovkin, M. A calmodulin binding protein from Arabidopsis is induced by ethylene and contains a DNA-binding motif. Biochem. Biophys. Res. Commun. 2000, 279, 762–769. [Google Scholar] [CrossRef] [PubMed]
- Yang, T.B.; Poovaiah, B.W. A calmodulin-binding/CGCG box DNA-binding protein family involved in multiple signaling pathways in plants. J. Boil. Chem. 2002, 277, 45049–45058. [Google Scholar] [CrossRef] [PubMed]
- Walley, J.W.; Coughlan, S.; Hudson, M.E.; Covington, M.F.; Kaspi, R.; Banu, G.; Harmer, S.L.; Dehesh, K. Mechanical stress induces biotic and abiotic stress responses via a novel cis-element. PLoS Genet. 2007, 3, e172. [Google Scholar] [CrossRef] [PubMed]
- Du, L.; Ali, G.S.; Simons, K.A.; Hou, J.; Yang, T.; Reddy, A.S.; Poovaiah, B.W. Ca(2+)/calmodulin regulates salicylic-acid-mediated plant immunity. Nature 2009, 457, 1154–1158. [Google Scholar] [CrossRef] [PubMed]
- Laluk, K.; Prasad, K.V.; Savchenko, T.; Celesnik, H.; Dehesh, K.; Levy, M.; Mitchell-Olds, T.; Reddy, A.S. The calmodulin-binding transcription factor SIGNAL RESPONSIVE1 is a novel regulator of glucosinolate metabolism and herbivory tolerance in Arabidopsis. Plant Cell Physiol. 2012, 53, 2008–2015. [Google Scholar] [CrossRef] [PubMed]
- Galon, Y.; Nave, R.; Boyce, J.M.; Nachmias, D.; Knight, M.R.; Fromm, H. Calmodulin-binding transcription activator (CAMTA) 3 mediates biotic defense responses in Arabidopsis. FEBS Lett. 2008, 582, 943–948. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mitsuda, N.; Isono, T.; Sato, M.H. Arabidopsis CAMTA family proteins enhance V-PPase expression in pollen. Plant Cell Physiol. 2003, 44, 975–981. [Google Scholar] [CrossRef] [PubMed]
- Babicki, S.; Arndt, D.; Marcu, A.; Liang, Y.; Grant, J.R.; Maciejewski, A.; Wishart, D.S. Heatmapper: Web-enabled heat mapping for all. Nucleic Acids Res. 2016, 44, W147–W153. [Google Scholar] [CrossRef] [PubMed]
- Tabas-Madrid, D.; Nogales-Cadenas, R.; Pascual-Montano, A. GeneCodis3: A non-redundant and modular enrichment analysis tool for functional genomics. Nucleic Acids Res. 2012, 40, W478–W483. [Google Scholar] [CrossRef] [PubMed]
- Jin, J.; Zhang, H.; Kong, L.; Gao, G.; Luo, J. PlantTFDB 3.0: A portal for the functional and evolutionary study of plant transcription factors. Nucleic Acids Res. 2014, 42, D1182–D1187. [Google Scholar] [CrossRef] [PubMed]
- Kankainen, M.; Holm, L. POBO, transcription factor binding site verification with bootstrapping. Nucleic Acids Res. 2004, 32, W222–W229. [Google Scholar] [CrossRef] [PubMed]










© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Prasad, K.V.S.K.; Xing, D.; Reddy, A.S.N. Vascular Plant One-Zinc-Finger (VOZ) Transcription Factors Are Positive Regulators of Salt Tolerance in Arabidopsis. Int. J. Mol. Sci. 2018, 19, 3731. https://doi.org/10.3390/ijms19123731
Prasad KVSK, Xing D, Reddy ASN. Vascular Plant One-Zinc-Finger (VOZ) Transcription Factors Are Positive Regulators of Salt Tolerance in Arabidopsis. International Journal of Molecular Sciences. 2018; 19(12):3731. https://doi.org/10.3390/ijms19123731
Chicago/Turabian StylePrasad, Kasavajhala V. S. K., Denghui Xing, and Anireddy S. N. Reddy. 2018. "Vascular Plant One-Zinc-Finger (VOZ) Transcription Factors Are Positive Regulators of Salt Tolerance in Arabidopsis" International Journal of Molecular Sciences 19, no. 12: 3731. https://doi.org/10.3390/ijms19123731
APA StylePrasad, K. V. S. K., Xing, D., & Reddy, A. S. N. (2018). Vascular Plant One-Zinc-Finger (VOZ) Transcription Factors Are Positive Regulators of Salt Tolerance in Arabidopsis. International Journal of Molecular Sciences, 19(12), 3731. https://doi.org/10.3390/ijms19123731