Heterologous Expression of Salvia miltiorrhiza MicroRNA408 Enhances Tolerance to Salt Stress in Nicotiana benthamiana
Abstract
:1. Introduction
2. Results
2.1. Sm-MIR408 Is Induced by Copper Deficiency and Salt Treatment
2.2. Heterologous Expression of Sm-MIR408 in Nicotiana benthamiana
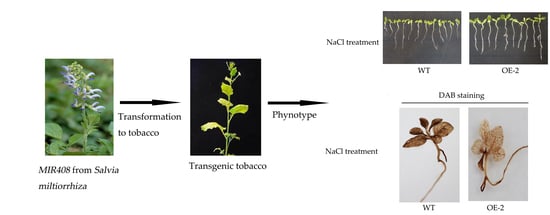
2.3. Sm-miR408 Confers Abiotic Stress Tolerance to Transgenic Plants
2.4. Overexpression of Sm-miR408 Reduced ROS Accumulation under Salt Stress
3. Discussion
4. Materials and Methods
4.1. Plant Material and Growth Conditions
4.2. Construction of Plant Expression Vector and Tobacco Transformation
4.3. Molecular Analysis of Transgenic Tobacco
4.4. Copper and Salt Treatment to S. miltiorrhiza and Tobacco
4.5. GUS Bioassays
4.6. Histochemical Detection of H2O2 and O2−
4.7. Determination of H2O2 Content
4.8. Measurement of SOD, POD, and CAT Activities
4.9. Expression Profile of ROS-Related Genes and Sm-MIR408 Response to Salt Stress
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Agarwal, P.; Reddy, M.P.; Chikara, J. Wrky: Its structure, evolutionary relationship, DNA-binding selectivity, role in stress tolerance and development of plants. Mol. Boil. Rep. 2011, 38, 3883–3896. [Google Scholar] [CrossRef]
- Yujia, L.; Xiaoyu, J.; Xianguang, N.; Min, Q.; Lei, Z.; Zilong, T.; Huimin, Z.; Lin, H.; Shengnan, L.; Bing, Z. Arabidopsis AtbHLH112 regulates the expression of genes involved in abiotic stress tolerance by binding to their E-box and GCG-box motifs. New Phytol. 2015, 207, 692–709. [Google Scholar]
- Altman, A. Recent advances in engineering plant tolerance to abiotic stress: Achievements and limitations. Curr. Opin. Biotechnol. 2005, 16, 123–132. [Google Scholar]
- Zhu, J.K. Salt and drought stress signal transduction in plants. Annu. Rev. Plant Boil. 2002, 53, 247–273. [Google Scholar] [CrossRef]
- Mittler, R.; Vanderauwera, S.; Gollery, M.; Van, B.F. Reactive oxygen gene network of plants. Trends Plant Sci. 2004, 9, 490–498. [Google Scholar] [CrossRef]
- Voinnet, O. Origin, biogenesis, and activity of plant micrornas. Cell 2009, 136, 669–687. [Google Scholar] [CrossRef]
- Chen, K.; Rajewsky, N. The evolution of gene regulation by transcription factors and microRNAs. Nat. Rev. Genet. 2007, 8, 93–103. [Google Scholar] [CrossRef]
- Hobert, O. Gene regulation by transcription factors and microRNAs. Science 2008, 319, 1785–1786. [Google Scholar] [CrossRef]
- Zhang, B. MicroRNA: A new target for improving plant tolerance to abiotic stress. J. Exp. Bot. 2015, 66, 1749–1761. [Google Scholar] [CrossRef]
- Palatnik, J.F.; Allen, E.; Wu, X.; Schommer, C.; Schwab, R.; Carrington, J.C.; Weigel, D. Control of leaf morphogenesis by microRNAs. Nature 2003, 425, 257–263. [Google Scholar] [CrossRef] [Green Version]
- Lauter, N.; Kampani, A.; Carlson, S.; Goebel, M.; Moose, S.P. microRNA172 down-regulates glossy15 to promote vegetative phase change in maize. Proc. Natl. Acad. Sci. USA 2005, 102, 9412–9417. [Google Scholar] [CrossRef]
- Yu, L.; Yu, X.; Shen, R.; He, Y. HYL1 gene maintains venation and polarity of leaves. Planta 2005, 221, 231–242. [Google Scholar] [CrossRef]
- Wu, G.; Park, M.Y.; Conway, S.R.; Wang, J.W.; Weigel, D.; Poethig, R.S. The sequential action of miR156 and miR172 regulates developmental timing in Arabidopsis. Cell 2009, 138, 750–759. [Google Scholar] [CrossRef]
- Zhao, L.; Kim, Y.; Dinh, T.T.; Chen, X. miR172 regulates stem cell fate and defines the inner boundary of APETALA3 and PISTILLATA expression domain in Arabidopsis floral meristems. Plant J. 2010, 51, 840–849. [Google Scholar] [CrossRef]
- Tao, W.; Sun, M.Y.; Wang, X.S.; Li, W.B.; Li, Y.G. Over-expression of GmGla-regulated soybean miR172a confers early flowering in transgenic Arabidopsis thaliana. Int. J. Mol. Sci. 2016, 17, 645. [Google Scholar]
- Reyes, J.L.; Chua, N.H. ABA induction of miR159 controls transcript levels of two MYB factors during Arabidopsis seed germination. Plant J. 2010, 49, 592–606. [Google Scholar] [CrossRef]
- Zhou, M.; Li, D.; Li, Z.; Hu, Q.; Yang, C.; Zhu, L.; Luo, H. Constitutive expression of a miR319 gene alters plant development and enhances salt and drought tolerance in transgenic creeping bentgrass. Plant Signal. Behav. 2014, 161, 1375–1391. [Google Scholar] [CrossRef]
- Sunkar, R.; Kapoor, A.; Zhu, J.K. Posttranscriptional induction of two Cu/Zn superoxide dismutase genes in Arabidopsis is mediated by downregulation of miR398 and important for oxidative stress tolerance. Plant Cell 2006, 18, 2051–2065. [Google Scholar] [CrossRef]
- Zhao, Q.; Nakashima, J.; Chen, F.; Yin, Y.; Fu, C.; Yun, J.; Shao, H.; Wang, X.; Wang, Z.Y.; Dixon, R.A. Laccase is necessary and nonredundant with peroxidase for lignin polymerization during vascular development in Arabidopsis. Plant Cell 2013, 25, 3976–3987. [Google Scholar] [CrossRef]
- Schuetz, M.; Benske, A.; Smith, R.A.; Watanabe, Y.; Tobimatsu, Y.; Ralph, J.; Demura, T.; Ellis, B.; Samuels, A.L. Laccases direct lignification in the discrete secondary cell wall domains of protoxylem. Plant Physiol. 2014, 166, 798–807. [Google Scholar] [CrossRef]
- Trindade, I.; Capitão, C.; Dalmay, T.; Fevereiro, M.P.; Santos, D.M.D. miR398 and miR408 are up-regulated in response to water deficit in Medicago truncatula. Planta 2010, 231, 705–716. [Google Scholar] [CrossRef]
- Barciszewskapacak, M.; Milanowska, K.; Knop, K.; Bielewicz, D.; Nuc, P.; Plewka, P.; Pacak, A.M.; Vazquez, F.; Karlowski, W.; Jarmolowski, A. Arabidopsis microRNA expression regulation in a wide range of abiotic stress responses. Front. Plant Sci. 2015, 6, 410. [Google Scholar]
- Zhou, L.; Liu, Y.; Liu, Z.; Kong, D.; Duan, M.; Luo, L. Genome-wide identification and analysis of drought-responsive micrornas in Oryza sativa. J. Exp. Bot. 2010, 61, 4157–4168. [Google Scholar] [CrossRef]
- Eldem, V.; Akcay, U.C.; Ozhuner, E.; Bakır, Y.; Uranbey, S.; Unver, T. Genome-wide identification of miRNAs responsive to drought in peach (Prunus persica) by high-throughput deep sequencing. PLoS ONE 2012, 7, e50298. [Google Scholar] [CrossRef]
- Zhang, H.; Zhao, X.; Li, J.; Cai, H.; Deng, X.W.; Li, L. microRNA408 is critical for the HY5-SPL7 gene network that mediates the coordinated response to light and copper. Plant Cell 2014, 26, 4933–4953. [Google Scholar] [CrossRef]
- Ma, C.; Burd, S.; Lers, A. MiR408 is involved in abiotic stress responses in Arabidopsis. Plant J. 2015, 84, 169–187. [Google Scholar] [CrossRef]
- Hajyzadeh, M.; Turktas, M.; Khawar, K.M.; Unver, T. MiR408 overexpression causes increased drought tolerance in chickpea. Gene 2015, 555, 186–193. [Google Scholar] [CrossRef]
- Committee for the Pharmacopoeia of People’s Republic of China. Pharmacopoeia of P.R. China; Part 1; Chemical Industry Publishing House: Biejing, China, 2015; p. 76. [Google Scholar]
- Hao, P.P.; Jiang, F.; Chen, Y.G.; Yang, J.; Zhang, K.; Zhang, M.X.; Zhang, C.; Zhao, Y.X.; Zhang, Y. Evidence for traditional chinese medication to treat cardiovascular disease. Nat. Rev. Cardiol. 2015, 12, 374. [Google Scholar] [CrossRef]
- Xu, X.; Jiang, Q.; Ma, X.; Ying, Q.; Shen, B.; Qian, Y.; Song, H.; Wang, H. Deep sequencing identifies tissue-specific microRNAs and their target genes involving in the biosynthesis of tanshinones in Salvia miltiorrhiza. PLoS ONE 2014, 9, e111679. [Google Scholar] [CrossRef]
- Zhang, L.; Wu, B.; Zhao, D.; Li, C.; Shao, F.; Lu, S. Genome-wide analysis and molecular dissection of the SPL gene family in Salvia miltiorrhiza. J. Integr. Plant Biol. 2014, 56, 38–50. [Google Scholar] [CrossRef]
- Guo, X.R.; Yang, X.B.; Wang, H.Q.; Ming, F.L.; She, X.; Cao, X.Y. Cloning and expression analysis of miR408 precursor sequences from Salvia miltiorrhiza. Plant Sci. J. 2016, 34, 430–438. [Google Scholar]
- Abdel-Ghany, S.E.; Pilon, M. MicroRNA-mediated systemic down-regulation of copper protein expression in response to low copper availability in Arabidopsis. J. Biol. Chem. 2008, 283, 15932–15945. [Google Scholar] [CrossRef]
- Zhang, H.; Li, L. SQUAMOSA promoter binding protein-like7 regulated microRNA408 is required for vegetative development in Arabidopsis. Plant J. 2013, 74, 98–109. [Google Scholar] [CrossRef]
- Baksa, I.; Nagy, T.; Barta, E.; Havelda, Z.; Várallyay, É.; Silhavy, D.; Burgyán, J.; Szittya, G. Identification of Nicotiana benthamiana microRNAs and their targets using high throughput sequencing and degradome analysis. BMC Genom. 2015, 16, 1025. [Google Scholar] [CrossRef]
- Yuan, S.; Li, Z.; Li, D.; Yuan, N.; Hu, Q.; Luo, H. Constitutive expression of rice microRNA528 alters plant development and enhances tolerance to salinity stress and nitrogen starvation in creeping bentgrass. Plant Physiol. 2015, 169, 576–593. [Google Scholar] [CrossRef]
- Luan, Y.; Cui, J.; Li, J.; Jiang, N.; Liu, P.; Meng, J. Effective enhancement of resistance to phytophthora infestans by overexpression of miR172a and b in Solanum lycopersicum. Planta 2017, 247, 127–138. [Google Scholar] [CrossRef]
- Yuan, N.; Yuan, S.; Li, Z.; Li, D.; Hu, Q.; Luo, H. Heterologous expression of a rice miR395 gene in Nicotiana tabacum impairs sulfate homeostasis. Sci. Rep. 2016, 6, 28791. [Google Scholar] [CrossRef]
- Zhao, J.; Yuan, S.; Zhou, M.; Yuan, N.; Li, Z.; Hu, Q.; Bethea, J.F.; Liu, H.; Li, S.; Luo, H. Transgenic creeping bentgrass overexpressing osa-miR393a exhibits altered plant development and improved multiple stress tolerance. Plant Biotechnol. J. 2018. [Google Scholar] [CrossRef]
- Zhou, H.; Hussain, S.S.; Hackenberg, M.; Bazanova, N.; Eini, O.; Li, J.; Gustafson, P.; Shi, B. Identification and characterisation of a previously unknown drought tolerance-associated microRNA in barley. Plant J. 2018, 95, 138–149. [Google Scholar] [CrossRef]
- Chen, L.; Zhang, X.; Liu, T.; Niu, S.; Wen, J.; Yi, B.; Ma, C.; Tu, J.; Fu, T. Identification of miRNAs that regulate silique development in Brassica napus. Plant Sci. 2018, 269, 106–117. [Google Scholar] [CrossRef]
- Zhao, X.; Hong, P.; Wu, J.; Chen, X.; Ye, X.; Pan, Y.; Wang, J.; Zhang, X. The tae-miR408-mediated control of TaTOC1 genes transcription is required for the regulation of heading time in wheat. Plant Physiol. 2016, 170, 1578. [Google Scholar] [CrossRef]
- Pan, J.; Huang, D.; Guo, Z.; Kuang, Z.; Zhang, H.; Xie, X.; Ma, Z.; Gao, S.; Lerdau, M.T.; Chu, C. Overexpression of microRNA408 enhances photosynthesis, growth, and seed yield in diverse plants. J. Integr. Plant Biol. 2018, 60, 323–340. [Google Scholar] [CrossRef]
- Zhang, J.P.; Yu, Y.; Feng, Y.Z.; Zhou, Y.F.; Zhang, F.; Yang, Y.W.; Lei, M.Q.; Zhang, Y.C.; Chen, Y.Q. miR408 regulates grain yield and photosynthesis via a phytocyanin protein. Plant Physiol. 2017, 175, 1175. [Google Scholar] [CrossRef]
- Song, Z.; Zhang, L.; Wang, Y.; Li, H.; Li, S.; Zhao, H.; Zhang, H. Constitutive expression of miR408 improves biomass and seed yield in Arabidopsis. Front. Plant Sci. 2017, 8, 2114. [Google Scholar] [CrossRef]
- Xie, F.; Wang, Q.; Sun, R.; Zhang, B. Deep sequencing reveals important roles of microRNAs in response to drought and salinity stress in cotton. J. Exp. Bot. 2015, 66, 789–804. [Google Scholar] [CrossRef]
- Anca, M.; Narendra, T. microRNAs targeting DEAD-box helicases are involved in salinity stress response in rice (Oryza sativa L.). BMC Plant Biol. 2012, 12, 183. [Google Scholar]
- Sun, X.; Liang, X.; Yan, W.; Yu, R.; Zhu, X.; Luo, X.; Gong, Y.; Wang, R.; Limera, C.; Zhang, K. Identification of novel and salt-responsive miRNAs to explore miRNA-mediated regulatory network of salt stress response in radish (Raphanus sativus L.). BMC Genom. 2015, 16, 197. [Google Scholar] [CrossRef]
- Wang, C.; Lu, G.; Hao, Y.; Guo, H.; Guo, Y.; Zhao, J.; Cheng, H. ABP9, a maize bZIP transcription factor, enhances tolerance to salt and drought in transgenic cotton. Planta 2017, 246, 453–469. [Google Scholar] [CrossRef]
- Taheri, P.; Kakooee, T. Reactive oxygen species accumulation and homeostasis are involved in plant immunity to an opportunistic fungal pathogen. J. Plant Physiol. 2017, 216, 152–163. [Google Scholar] [CrossRef]
- Yujie, F.; Kaifeng, L.; Hao, D.; Yan, X.; Huazhi, S.; Xianghua, L.; Lizhong, X. A stress-responsive NAC transcription factor SNAC3 confers heat and drought tolerance through modulation of reactive oxygen species in rice. J. Exp. Bot. 2015, 66, 6803–6817. [Google Scholar] [Green Version]
- Swain, D.M.; Sahoo, R.K.; Srivastava, V.K.; Tripathy, B.C.; Tuteja, R.; Tuteja, N. Function of heterotrimeric G-protein γ subunit RGG1 in providing salinity stress tolerance in rice by elevating detoxification of ROS. Planta 2016, 245, 367–383. [Google Scholar] [CrossRef]
- Mittler, R. Abiotic stress, the field environment and stress combination. Trends Plant Sci. 2006, 11, 15–19. [Google Scholar] [CrossRef]
- Zhang, S.; Zhuang, K.; Wang, S.; Lv, J.; Ma, N.N.; Meng, Q. A novel tomato SUME E3 ligase, SISIZ1, confers drought tolerance in transgenic tobacco. J. Integr. Plant Biol. 2017, 59, 102–117. [Google Scholar] [CrossRef]
- Lu, X.; Dun, H.; Lian, C.; Zhang, X.; Yin, W.; Xia, X. The role of peu-miR164 and its target PeNAC genes in response to abiotic stress in Populus euphratica. Plant Physiol. Biochem. 2017, 115, 418–438. [Google Scholar] [CrossRef]
- Tapken, W.; Ravet, K.; Pilon, M. Plastocyanin controls the stabilization of the thylakoid Cu-transporting P-type ATPase PAA2/HMA8 in response to low copper in Arabidopsis. J. Biol. Chem. 2012, 287, 18544–18550. [Google Scholar] [CrossRef]
- Ma, H.; Zhao, H.; Liu, Z.; Zhao, J. The phytocyanin gene family in rice (Oryza sativa L.): Genome-wide identification, classification and transcriptional analysis. PLoS ONE 2011, 6, e25184. [Google Scholar] [CrossRef]
- Ravet, K.; Pilon, M. Copper and iron homeostasis in plants: The challenges of oxidative stress. Antioxid. Redox Signal. 2013, 19, 919–932. [Google Scholar] [CrossRef]
- Jefferson, R.A.; Kavanagh, T.A.; Bevan, M.W. GUS fusions: Beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 1987, 6, 3901–3907. [Google Scholar] [CrossRef]




| Primer | Oligo Sequence 5′ to 3′ |
|---|---|
| SmUbi-F | ACCCTCACGGGGAAGACCATC |
| SmUbi-R | ACCACGGAGACGGAGGACAAG |
| MIR408-F | ACAGAAAATGGAGGCGAAGAAG |
| MIR408-R | GTCCCTAATCAGTGAGAGACACAGTAA |
| P-MIR408-F | ACGAGCACCGACTCTGATCATTG |
| P-MIR408-R | TCTTCTTCGCCTCCATTT TCTGTAT |
| 35S-F | ACAAAGGCGGCAACAAACG |
| 35S-R | GCCAGTCTTCACGGCGAGT |
| RT-MIR408-F | ACGGGGACGAGACAGAGCAT |
| RT-MIR408-R | GGCTTTCACACCAGCAACATAG |
| NbActin-F | CGTTATGGTTGGAATGGGACAGAA |
| NbActin-R | AAGAACAGGGTGCTCCTCGTGG |
| Copper-transporting ATPase PAA2-F | AAGAGGTCATCGGGGTTAGGA |
| Copper-transporting ATPase PAA2-R | GTAGCGTGAGTTCCACAGCATAAAG |
| Uclacyanin-2-F | ACTTGACGCCTCCGACCACT |
| Uclacyanin-2-R | ACTGCCTCTTCCCTAGACCATG |
| NbSOD-F | GGAGAGCCTTGTCTGATGG |
| NbSOD-R | TGGGTCCTGATTAGCAGTGGT |
| NbPOD-F | GTTGAGAGTTCTTGTCCTGGTGTT |
| NbPOD-R | TATTGGCTCCTCCCTGGTTTG |
| NbCAT-F | CACAGCCACGCTACTCAAGAC |
| NbCAT-R | CCACCCACCGACGAATAAAG |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Guo, X.; Niu, J.; Cao, X. Heterologous Expression of Salvia miltiorrhiza MicroRNA408 Enhances Tolerance to Salt Stress in Nicotiana benthamiana. Int. J. Mol. Sci. 2018, 19, 3985. https://doi.org/10.3390/ijms19123985
Guo X, Niu J, Cao X. Heterologous Expression of Salvia miltiorrhiza MicroRNA408 Enhances Tolerance to Salt Stress in Nicotiana benthamiana. International Journal of Molecular Sciences. 2018; 19(12):3985. https://doi.org/10.3390/ijms19123985
Chicago/Turabian StyleGuo, Xiaorong, Junfeng Niu, and Xiaoyan Cao. 2018. "Heterologous Expression of Salvia miltiorrhiza MicroRNA408 Enhances Tolerance to Salt Stress in Nicotiana benthamiana" International Journal of Molecular Sciences 19, no. 12: 3985. https://doi.org/10.3390/ijms19123985
APA StyleGuo, X., Niu, J., & Cao, X. (2018). Heterologous Expression of Salvia miltiorrhiza MicroRNA408 Enhances Tolerance to Salt Stress in Nicotiana benthamiana. International Journal of Molecular Sciences, 19(12), 3985. https://doi.org/10.3390/ijms19123985