Comparative Digital Gene Expression Analysis of Tissue-Cultured Plantlets of Highly Resistant and Susceptible Banana Cultivars in Response to Fusarium oxysporum
Abstract
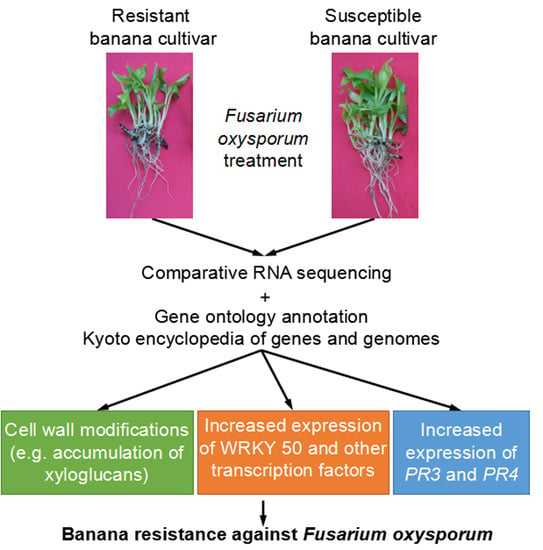
:1. Introduction
2. Results
2.1. Histological Observation of Fusarium Spreading in Root Tissues of Banana Cultivars
2.2. The Quality of Digital Gene Expression (DGE) Data
2.3. Differential Expression of Genes in the Roots of Banana Cultivars Resistant or Susceptible to Foc4
2.4. Gene Ontology (GO) Annotation Items Involved in the Resistance of Banana to Foc4
2.5. Kyoto Encyclopedia of Genes and Genomes (KEGG) Analysis
2.6. Changes in Thespatio-Temporal Distribution of Xyloglucan in Foc-Infected Banana
2.7. Quantitative Real-Time PCR (qRT-PCR) Validation of DGE Data
3. Discussion
3.1. Receptor Like Kinases
3.2. Trancription Factors
3.3. Canonical Defense Related and Cell Wall Associated Genes
3.4. Secondary Metabolism
3.5. New Genes with Putative Role in Foc4 Resistance
3.6. Comparative GO and KEGG Analysis and Screening of Potential Resistant Genes
4. Materials and Methods
4.1. Materials
4.2. Inoculation of Banana Cultivars with Pathogen
4.3. Immunolabelling of Xyloglucan
4.4. Observation of Pathogen Diffusion in Root Tissues of Banana
4.5. DGE Analysis
4.6. GO and KEGG Enrichment Analysis of Differentially Expressed Genes
4.7. qRT-PCR Validation Result
4.8. Statistical Analysis
5. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
Abbreviations
| BP | Biological process |
| BSA | Bovine serum albumin |
| CC | Cellular compartment |
| DEG | Differentially expressed genes |
| DGE | Digital gene expression |
| FDR | False discovery rate |
| Foc | Fusarium oxysporum f. sp. cubense |
| Foc4 | Race 4 of Foc |
| GO | Gene ontology |
| KEGG | Kyoto encyclopedia of genes and genomes |
| LOB | Lateral organ boundaries |
| LRR | Leucine-rich repeat |
| MF | Molecular function |
| NAC | NAM, ATAF1/2, CUC2 |
| PBS | Phosphate-buffered saline |
| PDA | Potato dextrose agar |
| PR | Pathogenesis-related |
| qRT-PCR | Quantitative real time PCR |
| RPKM | Reads per kilobase of transcript per million mapped reads |
| TFs | Transcription factors |
| UDP | Uridine diphosphate |
References
- FAOSTAT. Available online: http://www.fao.org/faostat/en/#data/QC/visualize (accessed on 20 January 2018).
- Hwang, S.C.; Ko, W.H. Cavendish banana cultivars resistant to Fusarium wilt acquired through somaclonal variation in Taiwan. Plant Dis. 2004, 88, 580–588. [Google Scholar] [CrossRef]
- Ploetz, R.C. Fusarium wilt of banana is caused by several pathogens referred to as Fusarium oxysporum f. sp. cubense. Phytopathology 2006, 96, 653–656. [Google Scholar] [CrossRef] [PubMed]
- Pegg, K.G.; Moore, N.Y.; Bentiey, S. Fusarium wilt of banana in Australia: A review. Aust. J. Agric. Res. 1996, 47, 637–650. [Google Scholar] [CrossRef]
- Swarupa, V.; Ravishankar, K.V.; Rekha, A. Plant defense response against Fusarium oxysporum and strategies to develop tolerant genotypes in banana. Planta 2014, 239, 735–751. [Google Scholar] [CrossRef] [PubMed]
- Li, C.Y.; Chen, S.; Zuo, C.W.; Sun, Q.M.; Ye, Q.; Yi, G.J.; Huang, B.H. The use of GFP-transformed isolates to study infection of banana with Fusarium oxysporum f. sp. cubense race 4. Eur. J. Plant Pathol. 2011, 131, 327–340. [Google Scholar] [CrossRef]
- Ma, L.; Jiang, S.; Lin, G.; Cai, J.; Ye, X.; Chen, H.; Li, M.; Li, H.; Takác, T.; Samaj, J.; et al. Wound-induced pectin methylesterases enhance banana (Musa spp. AAA) susceptibility to Fusarium oxysporum f. sp. cubense. J. Exp. Bot. 2013, 64, 2219–2229. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.L.; Fan, W.; Li, X.; Chen, H.B.; Takac, T.; Samajova, O.; Fabrice, M.R.; Xie, L.; Ma, J.; Samaj, J.; et al. Expression and distribution of extensins and AGPs in susceptible and resistant banana cultivars in response to wounding and Fusarium oxysporum. Sci. Rep. 2017, 7, 42400. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Yi, G.; Peng, X.; Huang, B.; Liu, E.; Zhang, J. Systemic acquired resistance in Cavendish banana induced by infection with an incompatible strain of Fusarium oxysporum f. sp. cubense. J. Plant Physiol. 2013, 170, 1039–1046. [Google Scholar] [CrossRef] [PubMed]
- Mahdavi, F.; Sariah, M.; Maziah, M. Expression of rice thaumatin-like protein gene in transgenic banana plants enhances resistance to Fusarium wilt. Appl. Biochem. Biotechnol. 2012, 166, 1008–1019. [Google Scholar] [CrossRef] [PubMed]
- Endah, R.; Beyene, G.; Kiggundu, A.; van den Berg, N.; Schlüter, U.; Kunert, K.; Chikwamba, R. Elicitor and Fusarium-induced expression of NPR1-like genes in banana. Plant Physiol. Biochem. 2008, 46, 1007–1014. [Google Scholar] [CrossRef] [PubMed]
- Luo, J.Y.; Pan, X.L.; Peng, T.C.; Chen, Y.Y.; Zhao, H.; Mu, L.; Peng, Y.; He, R.; Tang, H. DNA methylation patterns of banana leaves in response to Fusarium oxysporum f. sp. cubense tropical race 4. J. Integr. Agric. 2016, 15, 2736–2744. [Google Scholar] [CrossRef]
- Guo, Y.; Qiu, C.S.; Long, S.H.; Chen, P.; Hao, D.M.; Preisner, M.; Wang, H.; Wang, Y.F. Digital gene expression profiling of flax (Linumusitatissimum L.) stem peel identifies genes enriched in fiber-bearing phloem tissue. Gene 2017, 626, 32–40. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.Q.; Niu, Z.B.; Bao, Y.G.; Tian, Q.J.; Wang, H.G.; Kong, L.R.; Feng, D.S. Transcriptome analysis of genes related to resistance against powdery mildew in wheat-Thinopyrum alien addition disomic line germplasm SN6306. Gene 2016, 590, 5–17. [Google Scholar] [CrossRef] [PubMed]
- Cao, K.; Li, H.Y.; Wang, Q.; Zhao, P.; Zhu, G.Y.; Fang, W.C.; Chen, C.W.; Wang, X.W.; Wang, L. Comparative transcriptome analysis of genes involved in the response of resistant and susceptible peach cultivars to nematode infection. Sci. Hortic. 2017, 215, 20–27. [Google Scholar] [CrossRef]
- Wang, Z.; Zhang, J.B.; Jia, C.H.; Liu, J.H.; Li, Y.Q.; Yin, X.M.; Xu, B.Y.; Jin, Z.Q. De novo characterization of the banana root transcriptome and analysis of gene expression under Fusarium oxysporum f. sp. cubense tropical race 4 infection. BMC Genom. 2012, 13, 65. [Google Scholar]
- Li, C.; Shao, J.; Wang, Y.; Li, W.; Guo, D.; Yan, B.; Xia, Y.; Peng, M. Analysis of banana transcriptome and global gene expression profiles in banana roots in response to infection by race 1 and tropical race 4 of Fusarium oxysporum f. sp. cubense. BMC Genom. 2013, 14, 851. [Google Scholar] [CrossRef] [PubMed]
- Li, C.Y.; Deng, G.M.; Yang, J.; Viljoen, A.; Jin, Y.; Kuang, R.B.; Zuo, C.W.; Lv, Z.C.; Yang, Q.S.; Sheng, O.; et al. Transcriptome profiling of resistant and susceptible Cavendish banana roots following inoculation with Fusarium oxysporum f. sp. cubense tropical race 4. BMC Genom. 2012, 13, 374. [Google Scholar]
- Bai, T.T.; Xie, W.B.; Zhou, P.P.; Wu, Z.L.; Xiao, W.C.; Zhou, L.; Sun, J.; Ruan, X.L.; Li, H.P.; Zhang, Z.G. Transcriptome and expression profile analysis of highly resistant and susceptible banana roots challenged with Fusarium oxysporum f. sp. cubense tropical race 4. PLoS ONE 2013, 8, e73945. [Google Scholar] [CrossRef]
- D’Hont, A.; Denoeud, F.; Aury, J.M.; Baurens, F.C.; Carreel, F.; Garsmeur, O.; Noei, B.; Bocs, S.; Droc, G.; Rouard, M.; et al. The banana (Musa acuminata) genome and the evolution of monocotyledonous plants. Nature 2012, 488, 213–217. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Smith, M.K.; Whiley, A.W.; Searle, C.; Langdon, P.W.; Schaffer, B.; Pegg, K.G. Micropropagated bananas are more susceptible to Fusarium wilt than plants grown from conventional material. Aust. J. Agric. Res. 1998, 49, 1133–1139. [Google Scholar]
- Afzal, A.Z.; Wood, A.J.; Lightfoot, D.A. Plant receptor-like serine threonine kinases: Roles in signaling and plant defense. Mol. Plant Microbe Interact. 2008, 21, 507–517. [Google Scholar] [CrossRef] [PubMed]
- Mendy, B.; Wang’ombe, M.W.; Radakovic, Z.S.; Holbein, J.; Ilyas, M.; Chopra, D.; Holton, N.; Zipfel, C.; Grundler, F.M.; Siddique, S. Arabidopsis leucine-rich repeat receptor-like kinase NILR1 is required for induction of innate immunity to parasitic nematodes. PLoS Pathog. 2017, 13, e1006284. [Google Scholar] [CrossRef] [PubMed]
- Borassi, C.; Sede, A.R.; Mecchia, M.A.; Salgado Salter, J.D.; Marzol, E.; Muschietti, J.P.; Estevez, J.M. An update on cell surface proteins containing extensin-motifs. J. Exp. Bot. 2016, 67, 477–487. [Google Scholar] [CrossRef] [PubMed]
- Jin, J.H.; Zhang, H.X.; Tan, J.Y.; Yan, M.J.; Li, D.W.; Khan, A.; Dong, Z.H. A new ethylene-responsive factor CAPTI1 gene of pepper (Capsicum annuum L.) involved in the regulation of defense response to Phytophthora capsici. Front. Plant Sci. 2016, 6, 1217. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.J.; Sonbol, F.M.; Huot, B.; Gu, Y.N.; Withers, J.; Mwimba, M.; Yao, J.; He, S.Y.; Dong, X. Salicylic acid receptors activate jasmonic acid signalling through a non-canonical pathways to promote effector-triggered immunity. Nat. Commun. 2016, 7, 13099. [Google Scholar] [CrossRef] [PubMed]
- Feng, H.; Duan, X.; Zhang, Q.; Li, X.; Wang, B.; Huang, L.; Wang, X.; Kang, Z. The target gene of tae-miR164, a novel NAC transcription factor from the NAM subfamily, negatively regulates resistance of wheat to stripe rust. Mol. Plant Pathol. 2014, 15, 284–296. [Google Scholar] [CrossRef] [PubMed]
- Chen, N.; Wu, S.; Fu, J.; Cao, B.; Lei, J.; Chen, C.; Jiang, J. Overexpression of the eggplant (Solanummelongena) NAC family transcription factor SmNAC suppresses resistance to bacterial wilt. Sci. Rep. 2016, 6, 31568. [Google Scholar] [CrossRef]
- Pandey, S.P.; Somssich, I.E. The role of WRKY transcription factors in plant immunity. Plant Physiol. 2009, 150, 1648–1655. [Google Scholar] [CrossRef] [PubMed]
- Gao, Q.M.; Venugopal, S.; Navarre, D.; Kachroo, A. Low oleic acid-derived repression of jasmonic acid-inducible defense responses requires the WRKY50 and WRKY51 proteins. Plant Physiol. 2011, 155, 464–476. [Google Scholar] [CrossRef] [PubMed]
- Thatcher, L.F.; Powell, J.J.; Aieken, E.A.; Kazan, K.; Manners, J.M. The lateral organ boundaries domain transcription factor LBD20 functions in Fusarium wilt susceptibility and jasmonate signaling in Arabidopsis. Plant Physiol. 2012, 160, 407–418. [Google Scholar] [CrossRef] [PubMed]
- Shahnejat-Bushehri, S.; Nobmann, B.; Devi Allu, A.; Balazadeh, S. JUB1 suppresses Pseudomonas syringae-induced defense responses through accumulation of DELLA proteins. Plant Signal. Behav. 2016, 11, e1181245. [Google Scholar] [CrossRef] [PubMed]
- Di, X.T.; Gomila, J.; Takken, F.L.W. Involvement of salicylic acid, ethylene and jasmonic acid signalling pathways in the susceptibility of tomato to Fusarium oxysporum. Mol. Plant Pathol. 2017, 18, 1024–1035. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Shi, C.; Mu, X.Y.; Liu, C.; Shi, K.; Zhu, W.J.; Yang, Q. Cloning and expression of a wild eggplant cytochrome P450 gene, StoCYP77A2, involved in plant resistance to Verticillium dahliae. Plant Biotechnol. Rep. 2015, 9, 167–177. [Google Scholar] [CrossRef]
- Sels, J.; Mathys, J.; DeConinck, B.M.; Cammue, B.P.; DeBolle, M.F. Plant pathogenesis-related (PR) proteins: A focus on PR peptides. Plant Physiol. Biochem. 2008, 46, 941–950. [Google Scholar] [CrossRef] [PubMed]
- Farrokhi, N.; Burton, R.A.; Brownfield, L.; Hrmova, M.; Wilson, S.M.; Bacic, A.; Fincher, G.B. Plant cell wall biosynthesis: Genetic, biochemical and functional genomics approaches to the identification of key genes. Plant Biotechnol. J. 2006, 4, 145–167. [Google Scholar] [CrossRef] [PubMed]
- Pusztahelyi, T.; Holb, I.J.; Pócsi, I. Secondary metabolites in fungus-plant interactions. Front. Plant Sci. 2015, 6, 573. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bhattacharya, A.; Khanale, V.; Char, B. Plant circadian rhythm in stress signaling. Ind. J. Plant Physiol. 2017, 22, 147–155. [Google Scholar] [CrossRef]
- Wang, W.; Barnaby, J.Y.; Tada, Y.; Li, H.; Tör, M.; Caldelari, D.; Lee, D.U.; Fu, X.D.; Dong, X. Timing of plant immune responses by a central circadian regulator. Nature 2011, 470, 110–114. [Google Scholar] [CrossRef] [PubMed]
- Fan, G.; Dong, Y.; Deng, M.; Zhao, Z.; Niu, S.; Xu, E. Plant-pathogen interaction, circadian rhythm, and hormone-related gene expression provide indicators of phytoplasma infection in Paulownia fortunei. Int. J. Mol. Sci. 2014, 15, 23141–23162. [Google Scholar] [CrossRef] [PubMed]
- Seifi, H.; De Vleesschauwer, D.; Aziz, A.; Höfte, M. Modulating plant primary amino acid metabolism as a necrotrophic virulence strategy: The immune-regulatory role of asparagine synthetase in Botrytis cinerea-tomato interaction. Plant Signal. Behav. 2014, 9, e27995. [Google Scholar] [CrossRef] [PubMed]
- Brauc, S.; De Vooght, E.; Claeys, M.; Höfte, M.; Angenon, G. Influence of over-expression of cytosolic aspartate amino transferase on amino acid metabolism and defence responses against Botrytis cinerea infection in Arabidopsis thaliana. J. Plant Physiol. 2011, 168, 1813–1819. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.B.; Feng, Q.R.; Xu, C.X.; He, R.X.; Li, J.G.; Wang, Z.H. Screening of banana clones for resistance to Fusarium wilt (Fusarium oxysporum f. sp. cubense). J. South China Agric. Univ. 2006, 27, 9–12. [Google Scholar]
- Murashige, T.; Skoog, F. A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiol. Plant. 1962, 15, 473–497. [Google Scholar] [CrossRef]
- Xu, C.X.; Takáč, T.; Burbach, C.; Menzel, D.; Šamaj, J. Developmental localization and the role of hydroxyproline rich glycoproteins during somatic embryogenesis of banana (Musa spp. AAA). BMC Plant Biol. 2011, 11, 38. [Google Scholar] [CrossRef] [PubMed]
- Vitha, S.; Baluška, F.; Braun, M.; Šamaj, J.; Volkmann, D.; Barlow, P.W. Comparison of cryofixation and aldehyde fixation for plant actin immunocytochemistry: Aldehydes do not destroy F-actin. Histochem. J. 2000, 32, 457–466. [Google Scholar] [CrossRef] [PubMed]
- Marcus, S.E.; Verhertbruggen, Y.; Hervé, C.; Ordaz-Ortiz, J.J.; Farkas, V.; Pedersen, H.L.; Willats, W.G.T.; Knox, J.P. Pectic homogalacturonan masks abundant sets of xyloglucan epitopes in plant cell walls. BMC Plant Biol. 2008, 8, 60. [Google Scholar] [CrossRef] [PubMed]
- Götz, S.; García-Gómez, J.M.; Terol, J.; Williams, T.D.; Nagaraj, S.H.; Nueda, M.J.; Robles, M.; Talón, M.; Dopazo, J.; Conesa, A. High-throughput functional annotation and data mining with the Blast2GO suite. Nucleic Acids Res. 2008, 36, 3420–3435. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2 [Delta] [Delta] CT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Audic, S.; Claverie, J.M. The significance of digital gene expression profiles. Genome Res. 1997, 7, 986–995. [Google Scholar] [CrossRef] [PubMed]
- Dale, J.; James, A.; Paul, J.Y.; Khanna, H.; Smith, M.; Peraza-Echeverria, S.; Garcia-Bastidas, F.; Kema, G.; Waterhouse, P.; Mengersen, K.; et al. Transgenic Cavendish bananas with resistance to Fusarium wilt tropical race 4. Nat. Commun. 2017, 8, 1496. [Google Scholar] [CrossRef] [PubMed]



| Gene ID | Gene Description |
|---|---|
| LOC103991268 | Probable WRKY transcription factor 50 |
| LOC103971627 | Phenylalanine ammonia-lyase-like |
| LOC104000780 | Probable LRR receptor-like serine/threonine-protein kinase At1g74360 |
| LOC103983205 | UDP-glycosyltransferase 73C3-like |
| LOC103980149 | LOB domain-containing protein 41-like |
| LOC103990974 | Transcription factor JUNGBRUNNEN 1-like |
| LOC103978674 | Luminal-binding protein 2 |
| LOC103984595 | Uncharacterized |
| LOC103984038 | α-carbonic anhydrase 7-like |
| LOC103968789 | Curcumin synthase 3-like |
| LOC103983427 | Proline-rich receptor-like protein kinase PERK3 |
| LOC103972968 | Uncharacterized |
| Gene ID | Gene Description |
|---|---|
| LOC103993913 | Uncharacterized |
| LOC103978702 | Chitinase 1-like (PR-3) |
| LOC104000237 | Momilactone A synthase-like |
| LOC103978704 | Chitinase 1-like (PR-3) |
| LOC103989437 | Protein GOS9-like |
| LOC103975319 | Uncharacterized protein At1g15400-like |
| LOC103969557 | Transmembrane protein 45A-like |
| LOC103970573 | β-glucosidase 32-like |
| LOC103992490 | NAC domain-containing protein 68-like |
| LOC103977235 | Phenylpropanoylacetyl-CoA synthase-like |
| LOC103989217 | Uncharacterized |
| LOC103989972 | PR-4-like |
| LOC103992834 | Putative calcium-transporting ATPase 13, plasma membrane-type |
| LOC103988092 | Scarecrow-like protein 23 |
| LOC103980421 | Uncharacterized |
| LOC103993506 | Uncharacterized |
| LOC103974823 | α-carbonic anhydrase 7-like |
| LOC103987744 | Cytochrome P450 CYP72A219-like |
| LOC103979138 | Cytochrome P450 71A1-like |
| LOC104000899 | Uncharacterized |
| LOC104000297 | Leucine-rich repeat receptor protein kinase EXS |
| LOC103974226 | Uncharacterized |
| LOC103977900 | Uncharacterized |
| LOC103989489 | Miraculin-like |
| LOC103983075 | Uncharacterized |
| LOC103976880 | Probable xyloglucan endotransglucosylase/hydrolase protein 23 |
| LOC103979054 | Calcium-binding protein PBP1-like |
| LOC103974727 | Probable small nuclear ribonucleoprotein G |
| LOC103992513 | Probable WRKY transcription factor 70 |
| LOC103992023 | RING-H2 finger protein ATL2-like |
| LOC103989914 | Hexose carrier protein HEX6 |
| LOC103991268 | Probable WRKY transcription factor 50 |
| LOC104000052 | Putative UDP-glucuronate:xylan α-glucuronosyltransferase 3 |
| GO ID | Term | Percentage of DEGs before Infection (%) | Percentage of DEGs after Infection (%) |
|---|---|---|---|
| GO:0015706 | Nitrate transport | 4.17 | 4.17 |
| GO:0006073 | Cellular glucan metabolic process | 7.69 | 7.69 |
| GO:0005975 | Carbohydrate metabolic process | 4.29 | 2.86 |
| GO:0010030 | Positive regulation of seed germination | 33.33 | 33.33 |
| GO:0080167 | Response to karrikin | 6.82 | 2.27 |
| GO:0009611 | Response to wounding | 7.04 | 1.41 |
| GO:0010167 | Response to nitrate | 3.57 | 3.57 |
| GO:0016998 | Cell wall macromolecule catabolic process | 16.67 | 16.67 |
| GO:0009408 | Response to heat | 1.82 | 3.64 |
| GO:0044264 | Cellular polysaccharide metabolic process | 25 | 25 |
| GO:0006508 | Proteolysis | 3.77 | 0.94 |
| GO:0009407 | Toxin catabolic process | 5.26 | 5.26 |
| GO:0050832 | Defense response to fungus | 5.13 | 5.13 |
| GO:0032259 | Methylation | 1.92 | 5.76 |
| GO:0010583 | Response to cyclopentenone | 10 | 10 |
| GO:0009409 | Response to cold | 1.08 | 1.08 |
| GO:0006032 | Chitin catabolic process | 16.67 | 16.67 |
| GO:0009736 | Cytokinin-activated signaling pathway | 20 | 20 |
| GO:0009505 | Plant-type cell wall | 5.56 | 4.17 |
| GO:0005618 | Cell wall | 1.34 | 0.67 |
| GO:0005887 | Integral component of plasma membrane | 11.11 | 11.11 |
| GO:0043231 | Intracellular membrane-bounded organelle | 4.83 | 1.61 |
| GO:0044444 | Cytoplasmic part | 3.13 | 3.13 |
| GO:0009506 | Plasmodesma | 0.47 | 0.94 |
| GO:0016020 | Membrane | 1.13 | 0.23 |
| GO:0005773 | Vacuole | 1.33 | 1.33 |
| GO:0003700 | Sequence-specific DNA binding transcription factor activity | 1.22 | 2.45 |
| GO:0008168 | Methyltransferase activity | 2.5 | 7.5 |
| GO:0016757 | Transferase activity, transferring glycosyl groups | 4.76 | 2.38 |
| GO:0004568 | Chitinase activity | 12.5 | 12.5 |
| GO:0004674 | Protein serine/threonine kinase activity | 0.63 | 0.63 |
| GO:0000166 | Nucleotide binding | 0.61 | 0.61 |
| GO:0004601 | Peroxidase activity | 4 | 4 |
| GO:0030599 | Pectinesterase activity | 10 | 10 |
| GO:0004722 | Protein serine/threonine phosphatase activity | 3.33 | 3.33 |
| GO:0016762 | Xyloglucan:xyloglucosyl transferase activity | 7.69 | 7.69 |
| GO:0004190 | Aspartic-type endopeptidase activity | 6.67 | 6.67 |
| Comparison Pair | GO Annotation | Gene ID | Gene Description |
|---|---|---|---|
| YKCK–YKT | RNA splicing | LOC103989140 | Uncharacterized |
| Cellular zinc ion homeostasis | LOC103984038 | α-carbonic anhydrase 7-like | |
| One-carbon metabolic process | |||
| Carbonate dehydratase activity | |||
| Calcium-transporting ATPase activity | |||
| BXCK–BXT | Anaerobic respiration | LOC103970727 | Kelch repeat-containing protein At3g27220-like |
| LOC103984439 | Uncharacterized | ||
| Iron ion binding | LOC103985201 | Cytochrome c-like | |
| LOC103996064 | Glutamate synthase 1 [NADH], chloroplastic-like | ||
| LOC103990398 | Abscisic acid 8′-hydroxylase 1-like | ||
| LOC103974663 | Trans-cinnamate 4-monooxygenase-like | ||
| LOC103972072 | Cytochrome P450 84A1-like | ||
| Heme binding | LOC103985201 | Cytochrome c-like | |
| LOC103990398 | Abscisic acid 8′-hydroxylase 1-like | ||
| LOC103974663 | Trans-cinnamate 4-monooxygenase-like | ||
| LOC103972072 | Cytochrome P450 84A1-like | ||
| Electron carrier activity | LOC103985201 | Cytochrome c-like | |
| LOC103990398 | Abscisic acid 8′-hydroxylase 1-like | ||
| LOC103974663 | Trans-cinnamate 4-monooxygenase-like | ||
| LOC103972072 | Cytochrome P450 84A1-like | ||
| BXT–YKT | DNA binding | LOC103972990 | Protein CUP-SHAPED COTYLEDON 3-like |
| LOC103984324 | Uncharacterized | ||
| LOC103982987 | Auxin-responsive protein IAA4-like | ||
| LOC103985820 | NAC domain-containing protein 21/22-like | ||
| LOC103990974 | TF JUNGBRUNNEN 1-like | ||
| LOC103975993 | Ethylene-responsive TF ERF107-like | ||
| LOC103993650 | Ethylene-responsive TF 11-like | ||
| LOC103992490 | NAC domain-containing protein 68-like | ||
| LOC103971736 | TF MYB6-like |
| KEGG ID | Term |
|---|---|
| mus00020 | Citrate cycle |
| mus00100 | Steroid biosynthesis |
| mus00260 | Glycine, serine and threonine metabolism |
| mus00280 | Valine, leucine and isoleucine degradation |
| mus00310 | Lysine degradation |
| mus00450 | Selenocompound metabolism |
| mus00710 | Carbon fixation in photosynthetic organisms |
| mus00750 | Vitamin B6 metabolism |
| mus03040 | Spliceosome |
| mus04145 | Phagosome |
| Comparison Pair | Pathway ID | Pathway | No. of DEGs | Up-Regulated DEGs | Down-Regulated DEGs |
|---|---|---|---|---|---|
| BXCK–YKCK | mus01110 | Biosynthesis of secondary metabolites | 52 | 36 | 16 |
| mus00940 | Phenylpropanoid biosynthesis | 19 | 13 | 6 | |
| mus00941 | Flavonoid biosynthesis | 9 | 9 | 0 | |
| mus00360 | Phenylalanine metabolism | 14 | 5 | 9 | |
| mus01100 | Metabolic pathways | 68 | 39 | 29 | |
| mus00960 | Tropane, piperidine and pyridine alkaloid biosynthesis | 5 | 4 | 1 | |
| mus00950 | Isoquinoline alkaloid biosynthesis | 4 | 3 | 1 | |
| mus00500 | Starch and sucrose metabolism | 12 | 4 | 8 | |
| mus00480 | Glutathione metabolism | 6 | 5 | 1 | |
| mus00100 | Steroid biosynthesis | 4 | 0 | 4 | |
| YKCK–YKT | mus04712 | Circadian rhythm—plant | 2 | 1 | 1 |
| BXCK–BXT | mus01100 | Metabolic pathways | 44 | 13 | 31 |
| mus01110 | Biosynthesis of secondary metabolites | 27 | 11 | 16 | |
| mus00430 | Taurine and hypotaurine metabolism | 4 | 0 | 4 | |
| mus00940 | Phenylpropanoid biosynthesis | 9 | 5 | 4 | |
| mus00010 | Glycolysis/Gluconeogenesis | 6 | 0 | 6 | |
| mus00500 | Starch and sucrose metabolism | 7 | 0 | 7 | |
| mus00360 | Phenylalanine metabolism | 5 | 3 | 2 | |
| mus00941 | Flavonoid biosynthesis | 3 | 2 | 1 | |
| mus00250 | Alanine, aspartate and glutamate metabolism | 3 | 0 | 3 | |
| mus00592 | α-Linolenic acid metabolism | 3 | 1 | 2 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Niu, Y.; Hu, B.; Li, X.; Chen, H.; Takáč, T.; Šamaj, J.; Xu, C. Comparative Digital Gene Expression Analysis of Tissue-Cultured Plantlets of Highly Resistant and Susceptible Banana Cultivars in Response to Fusarium oxysporum. Int. J. Mol. Sci. 2018, 19, 350. https://doi.org/10.3390/ijms19020350
Niu Y, Hu B, Li X, Chen H, Takáč T, Šamaj J, Xu C. Comparative Digital Gene Expression Analysis of Tissue-Cultured Plantlets of Highly Resistant and Susceptible Banana Cultivars in Response to Fusarium oxysporum. International Journal of Molecular Sciences. 2018; 19(2):350. https://doi.org/10.3390/ijms19020350
Chicago/Turabian StyleNiu, Yuqing, Bei Hu, Xiaoquan Li, Houbin Chen, Tomáš Takáč, Jozef Šamaj, and Chunxiang Xu. 2018. "Comparative Digital Gene Expression Analysis of Tissue-Cultured Plantlets of Highly Resistant and Susceptible Banana Cultivars in Response to Fusarium oxysporum" International Journal of Molecular Sciences 19, no. 2: 350. https://doi.org/10.3390/ijms19020350
APA StyleNiu, Y., Hu, B., Li, X., Chen, H., Takáč, T., Šamaj, J., & Xu, C. (2018). Comparative Digital Gene Expression Analysis of Tissue-Cultured Plantlets of Highly Resistant and Susceptible Banana Cultivars in Response to Fusarium oxysporum. International Journal of Molecular Sciences, 19(2), 350. https://doi.org/10.3390/ijms19020350