A Phenotyping Method of Giant Cells from Root-Knot Nematode Feeding Sites by Confocal Microscopy Highlights a Role for CHITINASE-LIKE 1 in Arabidopsis
Abstract
:1. Introduction
2. Results and Discussion
2.1. Galls and GCs Induced by M. javanica in Arabidopsis and Cucumber Are Neatly Observed under Confocal Microscopy at Any Stage of Development
2.2. 3D Reconstruction of Galls and GCs Induced by M. javanica in Arabidopsis and Cucumber after Optical Sectioning of Fixed and Cleared Galls
2.3. Assigning a Role for CTL1 in RKN Feeding Sites by Confocal Based Quantification of GCs’ Size
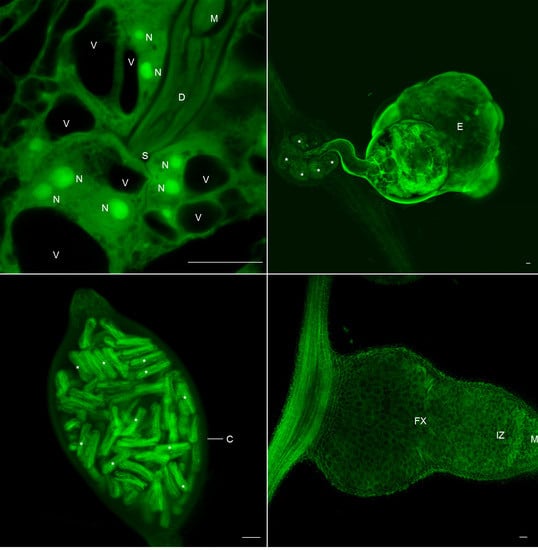
2.4. Clearing and Visualization of Hidden Structures during the Nematode and Other Biotic Interactions
3. Materials and Methods
3.1. Plant Material, Nematode Population Inoculation, and Infection
3.2. Fixation and Clearing of the Biological Samples
3.3. Confocal Microscopy
3.4. Image Processing
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Escobar, C.; Barcala, M.; Cabrera, J.; Fenoll, C. Overview of root-knot nematodes and giant cells. Adv. Bot. Res. 2015, 73, 1–32. [Google Scholar]
- Sobczak, M.; Golinowski, W. Structure of cyst nematode feeding sites. In Cell Biology of Plant Nematode Parasitism; Berg, R.H., Taylor, C.G., Eds.; Springer: Berlin/Heidelberg, Germany, 2009; pp. 153–187. [Google Scholar]
- Cabrera, J.; Diaz-Manzano, F.E.; Barcala, M.; Arganda-Carreras, I.; de Almeida-Engler, J.; Engler, G.; Fenoll, C.; Escobar, C. Phenotyping nematode feeding sites: Three-dimensional reconstruction and volumetric measurements of giant cells induced by root-knot nematodes in arabidopsis. New Phytol. 2015, 206, 868–880. [Google Scholar] [CrossRef] [PubMed]
- Dropkin, V. Pathology of meloidogyne—Galling, giant cell formation, effects on host physiology. EPPO Bull. 1972, 2, 23–32. [Google Scholar] [CrossRef]
- Wyss, U.; Grundler, F.M.W.; Munch, A. The parasitic behaviour of second-stage juveniles of Meloidogyne incognita in roots of Arabidopsis thaliana. Nematologica 1992, 38, 98–111. [Google Scholar] [CrossRef]
- Wieczorek, K. Cell wall alterations in nematode-infected roots. Adv. Bot. Res. 2015, 73, 61–90. [Google Scholar]
- De Almeida Engler, J.; Vieira, P.; Rodiuc, N.; Grossi de Sa, M.F.; Engler, G. The plant cell cycle machinery: Usurped and modulated by plant-parasitic nematodes. Adv. Bot. Res. 2015, 73, 91–118. [Google Scholar]
- Miyara, S.B.; Ionit, I.; Buki, P.; Kolomiets, M. Chapter six-the role of lipid signalling in regulating plant-nematode interactions. Adv. Bot. Res. 2015, 73, 139–166. [Google Scholar]
- Siddique, S.; Grundler, F.M.W. Metabolism in nematode feeding sites. Adv. Bot. Res. 2015, 73, 119–138. [Google Scholar]
- Cabrera, J.; Díaz-Manzano, F.E.; Fenoll, C.; Escobar, C. Developmental pathways mediated by hormones in nematode feeding sites. Adv. Bot. Res. 2015, 73, 167–188. [Google Scholar]
- Fester, T.; Berg, R.H.; Taylor, C.G. An easy method using glutaraldehyde-introduced fluorescence for the microscopic analysis of plant biotrophic interactions. J. Microsc. 2008, 231, 342–348. [Google Scholar] [CrossRef] [PubMed]
- De Souza, A.J.D., Jr.; Pierre, O.; Coelho, R.R.; Grossi-de-Sa, M.F.; Engler, G.; de Almeida Engler, J. Application of nuclear volume measurements to comprehend the cell cycle in root-knot nematode-induced giant cells. Front. Plant Sci. 2017, 8, 961. [Google Scholar]
- Ohtsu, M.; Sato, Y.; Kurihara, D.; Suzaki, T.; Kawaguchi, M.; Maruyama, D.; Higashiyama, T. Spatiotemporal deep imaging of syncytium induced by the soybean cyst nematode Heterodera glycines. Protoplasma 2017, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Vieira, P.; Engler, G.; de Almeida Engler, J. Whole-mount confocal imaging of nuclei in giant feeding cells induced by root-knot nematodes in arabidopsis. New Phytol. 2012, 195, 488–496. [Google Scholar] [CrossRef] [PubMed]
- Hasegawa, J.; Sakamoto, Y.; Nakagami, S.; Aida, M.; Sawa, S.; Matsunaga, S. Three-dimensional imaging of plant organs using a simple and rapid transparency technique. Plant Cell Physiol. 2016, 57, 462–472. [Google Scholar] [CrossRef] [PubMed]
- Migneault, I.; Dartiguenave, C.; Bertrand, M.J.; Waldron, K.C. Glutaraldehyde: Behavior in aqueous solution, reaction with proteins, and application to enzyme crosslinking. Biotechniques 2004, 37, 790–806. [Google Scholar] [PubMed]
- Lee, K.; Choi, S.; Yang, C.; Wu, H.-C.; Yu, J. Autofluorescence generation and elimination: A lesson from glutaraldehyde. Chem. Commun. 2013, 49, 3028–3030. [Google Scholar] [CrossRef] [PubMed]
- Susaki, E.A.; Ueda, H.R. Whole-body and whole-organ clearing and imaging techniques with single-cell resolution: Toward organism-level systems biology in mammals. Cell Chem. Biol. 2016, 23, 137–157. [Google Scholar] [CrossRef] [PubMed]
- Olmo, R.; Cabrera, J.; Moreno-Risueno, M.A.; Fukaki, H.; Fenoll, C.; Escobar, C. Molecular transducers from roots are triggered in arabidopsis leaves by root-knot nematodes for successful feeding site formation: A conserved post-embryogenic de novo organogenesis program? Front. Plant Sci. 2017, 8. [Google Scholar] [CrossRef] [PubMed]
- Hermans, C.; Porco, S.; Vandenbussche, F.; Gille, S.; De Pessemier, J.; Van Der Straeten, D.; Verbruggen, N.; Bush, D.R. Dissecting the role of chitinase-like1 in nitrate-dependent changes in root architecture. Plant Physiol. 2011, 157, 1313–1326. [Google Scholar] [CrossRef] [PubMed]
- Hermans, C.; Porco, S.; Verbruggen, N.; Bush, D.R. Chitinase-like protein ctl1 plays a role in altering root system architecture in response to multiple environmental conditions. Plant Physiol. 2010, 152, 904–917. [Google Scholar] [CrossRef] [PubMed]
- Barcala, M.; Garcia, A.; Cabrera, J.; Casson, S.; Lindsey, K.; Favery, B.; Garcia-Casado, G.; Solano, R.; Fenoll, C.; Escobar, C. Early transcriptomic events in microdissected arabidopsis nematode-induced giant cells. Plant J. Cell Mol. Biol. 2010, 61, 698–712. [Google Scholar] [CrossRef] [PubMed]
- Portillo, M.; Cabrera, J.; Lindsey, K.; Topping, J.; Andres, M.F.; Emiliozzi, M.; Oliveros, J.C.; Garcia-Casado, G.; Solano, R.; Koltai, H.; et al. Distinct and conserved transcriptomic changes during nematode-induced giant cell development in tomato compared with arabidopsis: A functional role for gene repression. New Phytol. 2013, 197, 1276–1290. [Google Scholar] [CrossRef] [PubMed]
- Cabrera, J.; Barcala, M.; García, A.; Rio-Machín, A.; Medina, C.; Jaubert-Possamai, S.; Favery, B.; Maizel, A.; Ruiz-Ferrer, V.; Fenoll, C.; et al. Differentially expressed small rnas in arabidopsis galls formed by Meloidogyne javanica: A functional role for mir390 and its tas3-derived tasirnas. New Phytol. 2016, 209, 1625–1640. [Google Scholar] [CrossRef] [PubMed]
- Borges, F.; Martienssen, R.A. The expanding world of small rnas in plants. Nat. Rev. Mol. Cell Biol. 2015, 16, 727–741. [Google Scholar] [CrossRef] [PubMed]
- Ruiz-Ferrer, V.; Voinnet, O. Roles of plant small rnas in biotic stress responses. Annu. Rev. Plant Biol. 2009, 60, 485–510. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.L.V.; Chekanova, J.A. Small rnas: Essential regulators of gene expression and defenses against environmental stresses in plants. Wiley Interdiscip. Rev. RNA 2016, 7, 356–381. [Google Scholar] [CrossRef] [PubMed]
- Cabrera, J.; Barcala, M.; Fenoll, C.; Escobar, C. The power of omics to identify plant susceptibility factors and to study resistance to root-knot nematodes. Curr. Issues Mol. Biol. 2016, 19, 53–72. [Google Scholar] [PubMed]
- Fosu-Nyarko, J.; Jones, M.G.; Wang, Z. Functional characterization of transcripts expressed in early-stage Meloidogyne javanica-induced giant cells isolated by laser microdissection. Mol. Plant Pathol. 2009, 10, 237–248. [Google Scholar] [CrossRef] [PubMed]
- Fuller, V.L.; Lilley, C.J.; Atkinson, H.J.; Urwin, P.E. Differential gene expression in arabidopsis following infection by plant-parasitic nematodes Meloidogyne incognita and Heterodera schachtii. Mol. Plant Pathol. 2007, 8, 595–609. [Google Scholar] [CrossRef] [PubMed]
- Hammes, U.Z.; Schachtman, D.P.; Berg, R.H.; Nielsen, E.; Koch, W.; McIntyre, L.M.; Taylor, C.G. Nematode-induced changes of transporter gene expression in arabidopsis roots. Mol. Plant-Microbe Interact. MPMI 2005, 18, 1247–1257. [Google Scholar] [CrossRef] [PubMed]
- Iberkleid, I.; Sela, N.; Miyara, S.B. Meloidogyne javanica fatty acid- and retinol-binding protein (mj-far-1) regulates expression of lipid-, cell wall-, stress- and phenylpropanoid-related genes during nematode infection of tomato. BMC Genom. 2015, 16, 272. [Google Scholar] [CrossRef] [PubMed]
- Jammes, F.; Lecomte, P.; de Almeida-Engler, J.; Bitton, F.; Martin-Magniette, M.L.; Renou, J.P.; Abad, P.; Favery, B. Genome-wide expression profiling of the host response to root-knot nematode infection in arabidopsis. Plant J. Cell Mol. Biol. 2005, 44, 447–458. [Google Scholar] [CrossRef] [PubMed]
- Kyndt, T.; Denil, S.; Haegeman, A.; Trooskens, G.; Bauters, L.; Van Criekinge, W.; De Meyer, T.; Gheysen, G. Transcriptional reprogramming by root knot and migratory nematode infection in rice. New Phytol. 2012, 196, 887–900. [Google Scholar] [CrossRef] [PubMed]
- Hussey, R.S.; Davis, E.L.; Baum, T.J. Secrets in secretions: Genes that control nematode parasitism of plants. Braz. J. Plant Physiol. 2002, 14, 183–194. [Google Scholar] [CrossRef]
- De Almeida Engler, J.; Gheysen, G. Nematode-induced endoreduplication in plant host cells: Why and how? Mol. Plant-Microbe Interact. 2012, 26, 17–24. [Google Scholar] [CrossRef] [PubMed]
- Cabrera, J.; Bustos, R.; Favery, B.; Fenoll, C.; Escobar, C. Nematic: A simple and versatile tool for the in silico analysis of plant-nematode interactions. Mol. Plant Pathol. 2014, 15, 627–636. [Google Scholar] [CrossRef] [PubMed]
- Sanchez-Rodriguez, C.; Bauer, S.; Hematy, K.; Saxe, F.; Ibanez, A.B.; Vodermaier, V.; Konlechner, C.; Sampathkumar, A.; Ruggeberg, M.; Aichinger, E.; et al. Chitinase-like1/pom-pom1 and its homolog ctl2 are glucan-interacting proteins important for cellulose biosynthesis in arabidopsis. Plant Cell 2012, 24, 589–607. [Google Scholar] [CrossRef] [PubMed]
- Cardona, A.; Saalfeld, S.; Schindelin, J.; Arganda-Carreras, I.; Preibisch, S.; Longair, M.; Tomancak, P.; Hartenstein, V.; Douglas, R.J. TrakEM2 software for neural circuit reconstruction. PLoS ONE 2012, 7, e38011. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jones, K.M.; Kobayashi, H.; Davies, B.W.; Taga, M.E.; Walker, G.C. How rhizobial symbionts invade plants: The sinorhizobium-medicago model. Nat. Rev. Microbiol. 2007, 5, 619–633. [Google Scholar] [CrossRef] [PubMed]
- Parniske, M. Intracellular accommodation of microbes by plants: A common developmental program for symbiosis and disease? Curr. Opin. Plant Biol. 2000, 3, 320–328. [Google Scholar] [CrossRef]
- Couzigou, J.-M.; Mondy, S.; Sahl, L.; Gourion, B.; Ratet, P. To be or noot to be: Evolutionary tinkering for symbiotic organ identity. Plant Signal. Behav. 2013, 8, 4498–4510. [Google Scholar] [CrossRef] [PubMed]
- Couzigou, J.-M.; Zhukov, V.; Mondy, S.; el Heba, G.A.; Cosson, V.; Ellis, T.N.; Ambrose, M.; Wen, J.; Tadege, M.; Tikhonovich, I.; et al. Nodule root and cochleata maintain nodule development and are legume orthologs of arabidopsis blade-on-petiole genes. Plant Cell 2012, 24, 4498–4510. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Franssen, H.J.; Xiao, T.T.; Kulikova, O.; Wan, X.; Bisseling, T.; Scheres, B.; Heidstra, R. Root developmental programs shape the Medicago truncatula nodule meristem. Development 2015, 142, 2941–2950. [Google Scholar] [CrossRef] [PubMed]
- Xiao, T.T.; Schilderink, S.; Moling, S.; Deinum, E.E.; Kondorosi, E.; Franssen, H.; Kulikova, O.; Niebel, A.; Bisseling, T. Fate map of Medicago truncatula root nodules. Development 2014, 141, 3517–3528. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bohlmann, H.; Wieczorek, K. Infection assay of cyst nematodes on arabidopsis roots. Bio-Protocol 2015, 5, e1596. [Google Scholar] [CrossRef] [PubMed]
- Díaz-Manzano, F.E.; Olmo, R.; Cabrera, J.; Barcala, M.; Escobar, C.; Fenoll, C. Long-term in vitro system for maintenance and amplification of root-knot nematodes in cucumis sativus roots. Front. Plant Sci. 2016, 7. [Google Scholar] [CrossRef] [PubMed]
- Fåhraeus, G. The infection of clover root hairs by nodule bacteria studied by a simple glass slide technique. J. Gen. Microbiol. 1957, 16, 374–381. [Google Scholar] [CrossRef] [PubMed]
- Schneider, C.A.; Rasband, W.S.; Eliceiri, K.W. Nih image to imagej: 25 years of image analysis. Nat. Methods 2012, 9, 671–675. [Google Scholar] [CrossRef] [PubMed]





© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cabrera, J.; Olmo, R.; Ruiz-Ferrer, V.; Abreu, I.; Hermans, C.; Martinez-Argudo, I.; Fenoll, C.; Escobar, C. A Phenotyping Method of Giant Cells from Root-Knot Nematode Feeding Sites by Confocal Microscopy Highlights a Role for CHITINASE-LIKE 1 in Arabidopsis. Int. J. Mol. Sci. 2018, 19, 429. https://doi.org/10.3390/ijms19020429
Cabrera J, Olmo R, Ruiz-Ferrer V, Abreu I, Hermans C, Martinez-Argudo I, Fenoll C, Escobar C. A Phenotyping Method of Giant Cells from Root-Knot Nematode Feeding Sites by Confocal Microscopy Highlights a Role for CHITINASE-LIKE 1 in Arabidopsis. International Journal of Molecular Sciences. 2018; 19(2):429. https://doi.org/10.3390/ijms19020429
Chicago/Turabian StyleCabrera, Javier, Rocio Olmo, Virginia Ruiz-Ferrer, Isidro Abreu, Christian Hermans, Isabel Martinez-Argudo, Carmen Fenoll, and Carolina Escobar. 2018. "A Phenotyping Method of Giant Cells from Root-Knot Nematode Feeding Sites by Confocal Microscopy Highlights a Role for CHITINASE-LIKE 1 in Arabidopsis" International Journal of Molecular Sciences 19, no. 2: 429. https://doi.org/10.3390/ijms19020429
APA StyleCabrera, J., Olmo, R., Ruiz-Ferrer, V., Abreu, I., Hermans, C., Martinez-Argudo, I., Fenoll, C., & Escobar, C. (2018). A Phenotyping Method of Giant Cells from Root-Knot Nematode Feeding Sites by Confocal Microscopy Highlights a Role for CHITINASE-LIKE 1 in Arabidopsis. International Journal of Molecular Sciences, 19(2), 429. https://doi.org/10.3390/ijms19020429