Sarcoma Spheroids and Organoids—Promising Tools in the Era of Personalized Medicine
Abstract
:1. Introduction
2. Three-Dimensional Models of Tumors
2.1. Tumor Spheroids
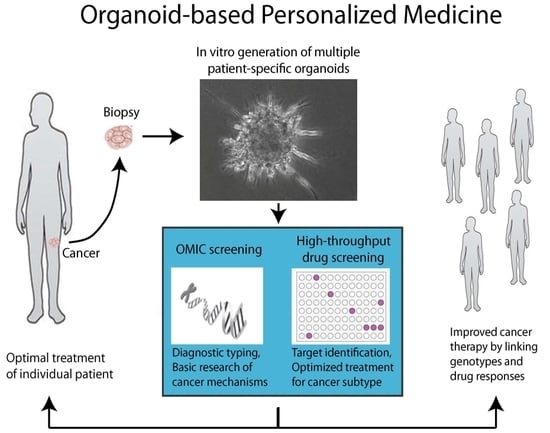
2.2. Tumor Organoids
3. Implementation of 3D Microenvironments (Hybrid Models for Investigating Angiogenesis, Influence of Immune Cells, and Tumor Dormancy)
4. Bone Tumor Niche
5. Applications for Sarcoma Biology Research
5.1. Functional Genomic Analysis
5.2. Preclinical Drug Screening
6. Clinical Opportunities
Acknowledgments
Conflicts of Interest
References
- Mohseny, A.B.; Hogendoorn, P.C. Concise review: Mesenchymal tumors: When stem cells go mad. Stem Cells 2011, 29, 397–403. [Google Scholar] [CrossRef] [PubMed]
- Stiller, C.A.; Trama, A.; Serraino, D.; Rossi, S.; Navarro, C.; Chirlaque, M.D.; Casali, P.G.; The RARECARE Working Group. Descriptive epidemiology of sarcomas in europe: Report from the rarecare project. Eur. J. Cancer 2013, 49, 684–695. [Google Scholar] [CrossRef] [PubMed]
- Gatta, G.; Ciccolallo, L.; Kunkler, I.; Capocaccia, R.; Berrino, F.; Coleman, M.P.; De Angelis, R.; Faivre, J.; Lutz, J.M.; Martinez, C.; et al. Survival from rare cancer in adults: a population-based study. Lancet Oncol. 2006, 7, 132–140. [Google Scholar] [CrossRef]
- Mastrangelo, G.; Coindre, J.M.; Ducimetiere, F.; Dei Tos, A.P.; Fadda, E.; Blay, J.Y.; Buja, A.; Fedeli, U.; Cegolon, L.; Frasson, A.; et al. Incidence of soft tissue sarcoma and beyond: A population-based prospective study in 3 european regions. Cancer 2012, 118, 5339–5348. [Google Scholar] [CrossRef] [PubMed]
- Dancsok, A.R.; Asleh-Aburaya, K.; Nielsen, T.O. Advances in sarcoma diagnostics and treatment. Oncotarget 2017, 8, 7068–7093. [Google Scholar] [CrossRef] [PubMed]
- Gerlinger, M.; Rowan, A.J.; Horswell, S.; Math, M.; Larkin, J.; Endesfelder, D.; Gronroos, E.; Martinez, P.; Matthews, N.; Stewart, A.; et al. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N. Engl. J. Med. 2012, 366, 883–892. [Google Scholar] [CrossRef] [PubMed]
- Hanahan, D.; Coussens, L.M. Accessories to the crime: Functions of cells recruited to the tumor microenvironment. Cancer Cell 2012, 21, 309–322. [Google Scholar] [CrossRef] [PubMed]
- Fazioli, F.; Colella, G.; Miceli, R.; Di Salvatore, M.G.; Gallo, M.; Boccella, S.; De Chiara, A.; Ruosi, C.; de Nigris, F. Post-surgery fluids promote transition of cancer stem cell-to-endothelial and akt/mtor activity, contributing to relapse of giant cell tumors of bone. Oncotarget 2017, 8, 85040–85053. [Google Scholar] [CrossRef] [PubMed]
- Salawu, A.; Fernando, M.; Hughes, D.; Reed, M.W.; Woll, P.; Greaves, C.; Day, C.; Alhajimohammed, M.; Sisley, K. Establishment and molecular characterisation of seven novel soft-tissue sarcoma cell lines. Br. J. Cancer 2016, 115, 1058–1068. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bruland, O.; Fodstad, O.; Pihl, A. The use of multicellular spheroids in establishing human sarcoma cell lines in vitro. Int. J. Cancer 1985, 35, 793–798. [Google Scholar] [CrossRef] [PubMed]
- Costa, E.C.; Gaspar, V.M.; Coutinho, P.; Correia, I.J. Optimization of liquid overlay technique to formulate heterogenic 3D co-cultures models. Biotechnol. Bioeng. 2014, 111, 1672–1685. [Google Scholar] [CrossRef] [PubMed]
- Ludwig, K.; Tse, E.S.; Wang, J.Y. Colon cancer cells adopt an invasive phenotype without mesenchymal transition in 3-D but not 2-D culture upon combined stimulation with EGF and crypt growth factors. BMC Cancer 2013, 13, 221. [Google Scholar] [CrossRef] [PubMed]
- Amann, A.; Zwierzina, M.; Gamerith, G.; Bitsche, M.; Huber, J.M.; Vogel, G.F.; Blumer, M.; Koeck, S.; Pechriggl, E.J.; Kelm, J.M.; et al. Development of an innovative 3d cell culture system to study tumour—Stroma interactions in non-small cell lung cancer cells. PLoS ONE 2014, 9, e92511. [Google Scholar] [CrossRef] [PubMed]
- Dufau, I.; Frongia, C.; Sicard, F.; Dedieu, L.; Cordelier, P.; Ausseil, F.; Ducommun, B.; Valette, A. Multicellular tumor spheroid model to evaluate spatio-temporal dynamics effect of chemotherapeutics: Application to the gemcitabine/CHK1 inhibitor combination in pancreatic cancer. BMC Cancer 2012, 12, 15. [Google Scholar] [CrossRef] [PubMed]
- Shankar, S.; Nall, D.; Tang, S.N.; Meeker, D.; Passarini, J.; Sharma, J.; Srivastava, R.K. Resveratrol inhibits pancreatic cancer stem cell characteristics in human and KrasG12D transgenic mice by inhibiting pluripotency maintaining factors and epithelial-mesenchymal transition. PLoS ONE 2011, 6, e16530. [Google Scholar] [CrossRef] [PubMed]
- Takagi, A.; Watanabe, M.; Ishii, Y.; Morita, J.; Hirokawa, Y.; Matsuzaki, T.; Shiraishi, T. Three-dimensional cellular spheroid formation provides human prostate tumor cells with tissue-like features. Anticancer Res. 2007, 27, 45–53. [Google Scholar] [PubMed]
- Clevers, H. Modeling development and disease with organoids. Cell 2016, 165, 1586–1597. [Google Scholar] [CrossRef] [PubMed]
- Lamichhane, S.P.; Arya, N.; Kohler, E.; Xiang, S.; Christensen, J.; Shastri, V.P. Recapitulating epithelial tumor microenvironment in vitro using three dimensional tri-culture of human epithelial, endothelial, and mesenchymal cells. BMC Cancer 2016, 16, 581. [Google Scholar] [CrossRef] [PubMed]
- Kolb, E.A.; Gorlick, R.; Maris, J.M.; Keir, S.T.; Morton, C.L.; Wu, J.; Wozniak, A.W.; Smith, M.A.; Houghton, P.J. Combination testing (stage 2) of the anti-IGF-1 receptor antibody IMC-A12 with rapamycin by the pediatric preclinical testing program. Pediatr. Blood Cancer 2012, 58, 729–735. [Google Scholar] [CrossRef] [PubMed]
- Hamilton, G. Multicellular spheroids as an in vitro tumor model. Cancer Lett. 1998, 131, 29–34. [Google Scholar] [CrossRef]
- Lawlor, E.R.; Scheel, C.; Irving, J.; Sorensen, P.H. Anchorage-independent multi-cellular spheroids as an in vitro model of growth signaling in Ewing tumors. Oncogene 2002, 21, 307–318. [Google Scholar] [CrossRef] [PubMed]
- Villaverde, M.S.; Gil-Cardeza, M.L.; Glikin, G.C.; Finocchiaro, L.M. Interferon-β lipofection I. Increased efficacy of chemotherapeutic drugs on human tumor cells derived monolayers and spheroids. Cancer Gene Ther. 2012, 19, 508–516. [Google Scholar] [CrossRef] [PubMed]
- Awad, O.; Yustein, J.T.; Shah, P.; Gul, N.; Katuri, V.; O’Neill, A.; Kong, Y.; Brown, M.L.; Toretsky, J.A.; Loeb, D.M. High ALDH activity identifies chemotherapy-resistant Ewing's sarcoma stem cells that retain sensitivity to EWS-FLI1 inhibition. PLoS ONE 2010, 5, e13943. [Google Scholar] [CrossRef] [PubMed]
- Zanoni, M.; Piccinini, F.; Arienti, C.; Zamagni, A.; Santi, S.; Polico, R.; Bevilacqua, A.; Tesei, A. 3D tumor spheroid models for in vitro therapeutic screening: A systematic approach to enhance the biological relevance of data obtained. Sci. Rep. 2016, 6, 19103. [Google Scholar] [CrossRef] [PubMed]
- Voissiere, A.; Jouberton, E.; Maubert, E.; Degoul, F.; Peyrode, C.; Chezal, J.M.; Miot-Noirault, E. Development and characterization of a human three-dimensional chondrosarcoma culture for in vitro drug testing. PLoS ONE 2017, 12, e0181340. [Google Scholar] [CrossRef] [PubMed]
- Strauss, S.J.; Ng, T.; Mendoza-Naranjo, A.; Whelan, J.; Sorensen, P.H. Understanding micrometastatic disease and anoikis resistance in Ewing family of tumors and osteosarcoma. Oncologist 2010, 15, 627–635. [Google Scholar] [CrossRef] [PubMed]
- Holmes, T.D.; El-Sherbiny, Y.M.; Davison, A.; Clough, S.L.; Blair, G.E.; Cook, G.P. A human NK cell activation/inhibition threshold allows small changes in the target cell surface phenotype to dramatically alter susceptibility to NK cells. J. Immunol. 2011, 186, 1538–1545. [Google Scholar] [CrossRef] [PubMed]
- Kailayangiri, S.; Altvater, B.; Meltzer, J.; Pscherer, S.; Luecke, A.; Dierkes, C.; Titze, U.; Leuchte, K.; Landmeier, S.; Hotfilder, M.; et al. The ganglioside antigen Gd2 is surface-expressed in Ewing sarcoma and allows for MHC-independent immune targeting. Br. J. Cancer 2012, 106, 1123–1133. [Google Scholar] [CrossRef] [PubMed]
- Kang, H.G.; Jenabi, J.M.; Zhang, J.; Keshelava, N.; Shimada, H.; May, W.A.; Ng, T.; Reynolds, C.P.; Triche, T.J.; Sorensen, P.H. E-cadherin cell-cell adhesion in ewing tumor cells mediates suppression of anoikis through activation of the ErbB4 tyrosine kinase. Cancer Res. 2007, 67, 3094–3105. [Google Scholar] [CrossRef] [PubMed]
- Graff, C.P.; Wittrup, K.D. Theoretical analysis of antibody targeting of tumor spheroids: Importance of dosage for penetration, and affinity for retention. Cancer Res. 2003, 63, 1288–1296. [Google Scholar] [PubMed]
- De Nigris, F.; Rossiello, R.; Schiano, C.; Arra, C.; Williams-Ignarro, S.; Barbieri, A.; Lanza, A.; Balestrieri, A.; Giuliano, M.T.; Ignarro, L.J.; et al. Deletion of Yin Yang 1 protein in osteosarcoma cells on cell invasion and CXCR4/angiogenesis and metastasis. Cancer Res. 2008, 68, 1797–1808. [Google Scholar] [CrossRef] [PubMed]
- De Nigris, F.; Crudele, V.; Giovane, A.; Casamassimi, A.; Giordano, A.; Garban, H.J.; Cacciatore, F.; Pentimalli, F.; Marquez-Garban, D.C.; Petrillo, A.; et al. CXCR4/YY1 inhibition impairs VEGF network and angiogenesis during malignancy. Proc. Natl. Acad. Sci. USA 2010, 107, 14484–14489. [Google Scholar] [CrossRef] [PubMed]
- Hassell, B.A.; Goyal, G.; Lee, E.; Sontheimer-Phelps, A.; Levy, O.; Chen, C.S.; Ingber, D.E. Human organ chip models recapitulate orthotopic lung cancer growth, therapeutic responses, and tumor dormancy in vitro. Cell. Rep. 2017, 21, 508–516. [Google Scholar] [CrossRef] [PubMed]
- Buschhaus, J.M.; Luker, K.E.; Luker, G.D. A facile, in vitro 384-well plate system to model disseminated tumor cells in the bone marrow microenvironment. Methods Mol. Biol. 2018, 1686, 201–213. [Google Scholar] [PubMed]
- Singh, M.; Mukundan, S.; Jaramillo, M.; Oesterreich, S.; Sant, S. Three-dimensional breast cancer models mimic hallmarks of size-induced tumor progression. Cancer Res. 2016, 76, 3732–3743. [Google Scholar] [CrossRef] [PubMed]
- Hu, G.; Li, D. Three-dimensional modeling of transport of nutrients for multicellular tumor spheroid culture in a microchannel. Biomed. Microdevices 2007, 9, 315–323. [Google Scholar] [CrossRef] [PubMed]
- Timmins, N.E.; Nielsen, L.K. Generation of multicellular tumor spheroids by the hanging-drop method. Methods Mol. Med. 2007, 140, 141–151. [Google Scholar] [PubMed]
- Kelm, J.M.; Timmins, N.E.; Brown, C.J.; Fussenegger, M.; Nielsen, L.K. Method for generation of homogeneous multicellular tumor spheroids applicable to a wide variety of cell types. Biotechnol. Bioeng. 2003, 83, 173–180. [Google Scholar] [CrossRef] [PubMed]
- Caplin, J.D.; Granados, N.G.; James, M.R.; Montazami, R.; Hashemi, N. Microfluidic organ-on-a-chip technology for advancement of drug development and toxicology. Adv. Healthc. Mater. 2015, 4, 1426–1450. [Google Scholar] [CrossRef] [PubMed]
- Friedrich, J.; Seidel, C.; Ebner, R.; Kunz-Schughart, L.A. Spheroid-based drug screen: Considerations and practical approach. Nat. Protoc. 2009, 4, 309–324. [Google Scholar] [CrossRef] [PubMed]
- Vinci, M.; Gowan, S.; Boxall, F.; Patterson, L.; Zimmermann, M.; Court, W.; Lomas, C.; Mendiola, M.; Hardisson, D.; Eccles, S.A. Advances in establishment and analysis of three-dimensional tumor spheroid-based functional assays for target validation and drug evaluation. BMC Biol. 2012, 10, 29. [Google Scholar] [CrossRef] [PubMed]
- Lovitt, C.J.; Shelper, T.B.; Avery, V.M. Advanced cell culture techniques for cancer drug discovery. Biology (Basel) 2014, 3, 345–367. [Google Scholar] [CrossRef] [PubMed]
- Fatehullah, A.; Tan, S.H.; Barker, N. Organoids as an in vitro model of human development and disease. Nat. Cell. Biol. 2016, 18, 246–254. [Google Scholar] [CrossRef] [PubMed]
- Gaebler, M.; Silvestri, A.; Haybaeck, J.; Reichardt, P.; Lowery, C.D.; Stancato, L.F.; Zybarth, G.; Regenbrecht, C.R.A. Three-dimensional patient-derived in vitro sarcoma models: Promising tools for improving clinical tumor management. Front. Oncol. 2017, 7, 203. [Google Scholar] [CrossRef] [PubMed]
- Stewart, E.; Federico, S.M.; Chen, X.; Shelat, A.A.; Bradley, C.; Gordon, B.; Karlstrom, A.; Twarog, N.R.; Clay, M.R.; Bahrami, A.; et al. Orthotopic patient-derived xenografts of paediatric solid tumours. Nature 2017, 549, 96–100. [Google Scholar] [CrossRef] [PubMed]
- Hong, A.L.; Tseng, Y.Y.; Cowley, G.S.; Jonas, O.; Cheah, J.H.; Kynnap, B.D.; Doshi, M.B.; Oh, C.; Meyer, S.C.; Church, A.J.; et al. Integrated genetic and pharmacologic interrogation of rare cancers. Nat. Commun. 2016, 7, 11987. [Google Scholar] [CrossRef] [PubMed]
- Gao, H.; Korn, J.M.; Ferretti, S.; Monahan, J.E.; Wang, Y.; Singh, M.; Zhang, C.; Schnell, C.; Yang, G.; Zhang, Y.; et al. High-throughput screening using patient-derived tumor xenografts to predict clinical trial drug response. Nat. Med. 2015, 21, 1318–1325. [Google Scholar] [CrossRef] [PubMed]
- Aparicio, S.; Hidalgo, M.; Kung, A.L. Examining the utility of patient-derived xenograft mouse models. Nat. Rev. Cancer 2015, 15, 311–316. [Google Scholar] [CrossRef] [PubMed]
- Reddy, K.; Zhou, Z.; Jia, S.F.; Lee, T.H.; Morales-Arias, J.; Cao, Y.; Kleinerman, E.S. Stromal cell-derived factor-1 stimulates vasculogenesis and enhances Ewing's sarcoma tumor growth in the absence of vascular endothelial growth factor. Int. J. Cancer 2008, 123, 831–837. [Google Scholar] [CrossRef] [PubMed]
- Lee, T.H.; Bolontrade, M.F.; Worth, L.L.; Guan, H.; Ellis, L.M.; Kleinerman, E.S. Production of VEGF165 by Ewing's sarcoma cells induces vasculogenesis and the incorporation of CD34+ stem cells into the expanding tumor vasculature. Int. J. Cancer 2006, 119, 839–846. [Google Scholar] [CrossRef] [PubMed]
- De Nigris, F.; Mancini, F.P.; Schiano, C.; Infante, T.; Zullo, A.; Minucci, P.B.; Al-Omran, M.; Giordano, A.; Napoli, C. Osteosarcoma cells induce endothelial cell proliferation during neo-angiogenesis. J. Cell. Physiol. 2013, 228, 846–852. [Google Scholar] [CrossRef] [PubMed]
- Howes, A.L.; Richardson, R.D.; Finlay, D.; Vuori, K. 3-dimensional culture systems for anti-cancer compound profiling and high-throughput screening reveal increases in EGFR inhibitor-mediated cytotoxicity compared to monolayer culture systems. PLoS One 2014, 9, e108283. [Google Scholar] [CrossRef] [PubMed]
- Shoval, H.; Karsch-Bluman, A.; Brill-Karniely, Y.; Stern, T.; Zamir, G.; Hubert, A.; Benny, O. Tumor cells and their crosstalk with endothelial cells in 3D spheroids. Sci. Rep. 2017, 7, 10428. [Google Scholar] [CrossRef] [PubMed]
- Chaddad, H.; Kuchler-Bopp, S.; Fuhrmann, G.; Gegout, H.; Ubeaud-Sequier, G.; Schwinte, P.; Bornert, F.; Benkirane-Jessel, N.; Idoux-Gillet, Y. Combining 2D angiogenesis and 3D osteosarcoma microtissues to improve vascularization. Exp. Cell. Res. 2017, 360, 138–145. [Google Scholar] [CrossRef] [PubMed]
- Kusumbe, A.P. Vascular niches for disseminated tumour cells in bone. J. Bone Oncol. 2016, 5, 112–116. [Google Scholar] [CrossRef] [PubMed]
- Augustine, T.N.; Dix-Peek, T.; Duarte, R.; Candy, G.P. Establishment of a heterotypic 3D culture system to evaluate the interaction of TREG lymphocytes and NK cells with breast cancer. J. Immunol. Methods 2015, 426, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Herter, S.; Morra, L.; Schlenker, R.; Sulcova, J.; Fahrni, L.; Waldhauer, I.; Lehmann, S.; Reislander, T.; Agarkova, I.; Kelm, J.M.; et al. A novel three-dimensional heterotypic spheroid model for the assessment of the activity of cancer immunotherapy agents. Cancer Immunol. Immunother. 2017, 66, 129–140. [Google Scholar] [CrossRef] [PubMed]
- Villasante, A.; Vunjak-Novakovic, G. Tissue-engineered models of human tumors for cancer research. Expert Opin. Drug Discov. 2015, 10, 257–268. [Google Scholar] [CrossRef] [PubMed]
- Tse, J.M.; Cheng, G.; Tyrrell, J.A.; Wilcox-Adelman, S.A.; Boucher, Y.; Jain, R.K.; Munn, L.L. Mechanical compression drives cancer cells toward invasive phenotype. Proc. Natl. Acad. Sci. USA 2012, 109, 911–916. [Google Scholar] [CrossRef] [PubMed]
- Gill, B.J.; West, J.L. Modeling the tumor extracellular matrix: Tissue engineering tools repurposed towards new frontiers in cancer biology. J. Biomech. 2014, 47, 1969–1978. [Google Scholar] [CrossRef] [PubMed]
- Rocco, G.; Martucci, N.; La Rocca, A.; La Manna, C.; De Luca, G.; Fazioli, F.; Mori, S. Postoperative local morbidity and the use of vacuum-assisted closure after complex chest wall reconstructions with new and conventional materials. Ann. Thorac. Surg. 2014, 98, 291–296. [Google Scholar] [CrossRef] [PubMed]
- Marturano-Kruik, A.; Villasante, A.; Vunjak-Novakovic, G. Bioengineered models of solid human tumors for cancer research. Methods Mol. Biol. 2016, 1502, 203–211. [Google Scholar] [PubMed]
- Fong, E.L.; Lamhamedi-Cherradi, S.E.; Burdett, E.; Ramamoorthy, V.; Lazar, A.J.; Kasper, F.K.; Farach-Carson, M.C.; Vishwamitra, D.; Demicco, E.G.; Menegaz, B.A.; et al. Modeling ewing sarcoma tumors in vitro with 3D scaffolds. Proc. Natl. Acad. Sci. USA 2013, 110, 6500–6505. [Google Scholar] [CrossRef] [PubMed]
- Infanger, D.W.; Lynch, M.E.; Fischbach, C. Engineered culture models for studies of tumor-microenvironment interactions. Annu. Rev. Biomed. Eng. 2013, 15, 29–53. [Google Scholar] [CrossRef] [PubMed]
- Santoro, M.; Lamhamedi-Cherradi, S.E.; Menegaz, B.A.; Ludwig, J.A.; Mikos, A.G. Flow perfusion effects on three-dimensional culture and drug sensitivity of Ewing sarcoma. Proc. Natl. Acad. Sci. USA 2015, 112, 10304–10309. [Google Scholar] [CrossRef] [PubMed]
- Lamhamedi-Cherradi, S.E.; Santoro, M.; Ramammoorthy, V.; Menegaz, B.A.; Bartholomeusz, G.; Iles, L.R.; Amin, H.M.; Livingston, J.A.; Mikos, A.G.; Ludwig, J.A. 3D tissue-engineered model of Ewing's sarcoma. Adv. Drug Deliv. Rev. 2014, 79–80, 155–171. [Google Scholar] [CrossRef] [PubMed]
- Dit Faute, M.A.; Laurent, L.; Ploton, D.; Poupon, M.F.; Jardillier, J.C.; Bobichon, H. Distinctive alterations of invasiveness, drug resistance and cell-cell organization in 3D-cultures of MCF-7, a human breast cancer cell line, and its multidrug resistant variant. Clin. Exp. Metastasis 2002, 19, 161–168. [Google Scholar] [CrossRef] [PubMed]
- Infante, T.; Mancini, F.P.; Lanza, A.; Soricelli, A.; de Nigris, F.; Napoli, C. Polycomb YY1 is a critical interface between epigenetic code and miRNA machinery after exposure to hypoxia in malignancy. Biochim. Biophys. Acta. 2015, 1853, 975–986. [Google Scholar] [CrossRef] [PubMed]
- Barretina, J.; Taylor, B.S.; Banerji, S.; Ramos, A.H.; Lagos-Quintana, M.; Decarolis, P.L.; Shah, K.; Socci, N.D.; Weir, B.A.; Ho, A.; et al. Subtype-specific genomic alterations define new targets for soft-tissue sarcoma therapy. Nat. Genet. 2010, 42, 715–721. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Qi, Y.; Hu, Y.; Yang, H.; Zhuang, R.; Hou, Y.; Tong, H.; Feng, Y.; Huang, Y.; Jiang, Q.; Ji, Q.; et al. Establishing a patient-derived xenograft model of human myxoid and round-cell liposarcoma. Oncotarget 2017, 8, 54320–54330. [Google Scholar] [CrossRef] [PubMed]
- Jour, G.; Scarborough, J.D.; Jones, R.L.; Loggers, E.; Pollack, S.M.; Pritchard, C.C.; Hoch, B.L. Molecular profiling of soft tissue sarcomas using next-generation sequencing: A pilot study toward precision therapeutics. Hum. Pathol. 2014, 45, 1563–1571. [Google Scholar] [CrossRef] [PubMed]
- Horman, S.R.; To, J.; Orth, A.P.; Slawny, N.; Cuddihy, M.J.; Caracino, D. High-content analysis of three-dimensional tumor spheroids: Investigating signaling pathways using small hairpin RNA. Nature Methods 2013, 10, v–vi. [Google Scholar] [CrossRef]
- Liu, T.; Shen, J.K.; Li, Z.; Choy, E.; Hornicek, F.J.; Duan, Z. Development and potential applications of CRISPR-Cas9 genome editing technology in sarcoma. Cancer Lett. 2016, 373, 109–118. [Google Scholar] [CrossRef] [PubMed]
- Hanes, R.; Grad, I.; Lorenz, S.; Stratford, E.W.; Munthe, E.; Reddy, C.C.; Meza-Zepeda, L.A.; Myklebost, O. Preclinical evaluation of potential therapeutic targets in dedifferentiated liposarcoma. Oncotarget 2016, 7, 54583–54595. [Google Scholar] [CrossRef] [PubMed]
- Drost, J.; van Jaarsveld, R.H.; Ponsioen, B.; Zimberlin, C.; van Boxtel, R.; Buijs, A.; Sachs, N.; Overmeer, R.M.; Offerhaus, G.J.; Begthel, H.; et al. Sequential cancer mutations in cultured human intestinal stem cells. Nature 2015, 521, 43–47. [Google Scholar] [CrossRef] [PubMed]
- Treindl, F.; Ruprecht, B.; Beiter, Y.; Schultz, S.; Dottinger, A.; Staebler, A.; Joos, T.O.; Kling, S.; Poetz, O.; Fehm, T.; et al. A bead-based western for high-throughput cellular signal transduction analyses. Nat. Commun. 2016, 7, 12852. [Google Scholar] [CrossRef] [PubMed]
- Kondo, T.; Kawaib, A. A proteomics approach for the development of sarcoma biomarkers. EuPA Open Proteom. 2014, 4, 121–128. [Google Scholar] [CrossRef]
- Oudar, O. Spheroids: Relation between tumour and endothelial cells. Crit. Rev. Oncol. Hematol. 2000, 36, 99–106. [Google Scholar] [CrossRef]
- Furbert-Harris, P.M.; Laniyan, I.; Harris, D.; Dunston, G.M.; Vaughn, T.; Abdelnaby, A.; Parish-Gause, D.; Oredipe, O.A. Activated eosinophils infiltrate MCF-7 breast multicellular tumor spheroids. Anticancer Res. 2003, 23, 71–78. [Google Scholar] [PubMed]
- Arai, K.; Sakamoto, R.; Kubota, D.; Kondo, T. Proteomic approach toward molecular backgrounds of drug resistance of osteosarcoma cells in spheroid culture system. Proteomics 2013, 13, 2351–2360. [Google Scholar] [CrossRef] [PubMed]
- Leon, I.E.; Cadavid-Vargas, J.F.; Resasco, A.; Maschi, F.; Ayala, M.A.; Carbone, C.; Etcheverry, S.B. In vitro and in vivo antitumor effects of the VO-chrysin complex on a new three-dimensional osteosarcoma spheroids model and a xenograft tumor in mice. J. Biol. Inorg. Chem. 2016, 21, 1009–1020. [Google Scholar] [CrossRef] [PubMed]
- Westhoff, M.A.; Fulda, S. Adhesion-mediated apoptosis resistance in cancer. Drug Resist. Updat. 2009, 12, 127–136. [Google Scholar] [CrossRef] [PubMed]
- McMillin, D.W.; Negri, J.M.; Mitsiades, C.S. The role of tumour-stromal interactions in modifying drug response: Challenges and opportunities. Nat. Rev. Drug Discov. 2013, 12, 217–228. [Google Scholar] [CrossRef] [PubMed]
- Benien, P.; Swami, A. 3D tumor models: History, advances and future perspectives. Future Oncol. 2014, 10, 1311–1327. [Google Scholar] [CrossRef] [PubMed]
- Hiscox, S.; Barrett-Lee, P.; Nicholson, R.I. Therapeutic targeting of tumor-stroma interactions. Expert Opin. Ther. Targets 2011, 15, 609–621. [Google Scholar] [CrossRef] [PubMed]
- Kunz-Schughart, L.A.; Freyer, J.P.; Hofstaedter, F.; Ebner, R. The use of 3-D cultures for high-throughput screening: The multicellular spheroid model. J. Biomol. Screen. 2004, 9, 273–285. [Google Scholar] [CrossRef] [PubMed]
- Walenta, S.; Doetsch, J.; Mueller-Klieser, W.; Kunz-Schughart, L.A. Metabolic imaging in multicellular spheroids of oncogene-transfected fibroblasts. J. Histochem. Cytochem. 2000, 48, 509–522. [Google Scholar] [CrossRef] [PubMed]
- Oloumi, A.; MacPhail, S.H.; Johnston, P.J.; Banath, J.P.; Olive, P.L. Changes in subcellular distribution of topoisomerase IIα correlate with etoposide resistance in multicell spheroids and xenograft tumors. Cancer Res. 2000, 60, 5747–5753. [Google Scholar] [PubMed]
- Pervez, S.; Kirkland, S.C.; Epenetos, A.A.; Mooi, W.J.; Evans, D.J.; Krausz, T. Effect of polarity and differentiation on antibody localization in multicellular tumour spheroid and xenograft models and its potential importance for in vivo immunotargeting. Int. J. Cancer 1989, 44, 940–947. [Google Scholar] [CrossRef] [PubMed]
- Roose, T.; Netti, P.A.; Munn, L.L.; Boucher, Y.; Jain, R.K. Solid stress generated by spheroid growth estimated using a linear poroelasticity model. Microvasc. Res. 2003, 66, 204–212. [Google Scholar] [CrossRef]
- Lichtenbeld, H.H.; Muller, A.D.; van Dam-Mieras, M.C.; Blijham, G.H. Tumor spheroid-induced vesicle formation on endothelial cells is associated with procoagulant properties. J. Cell. Sci. 1993, 106 Pt 2, 657–662. [Google Scholar] [PubMed]
- Konur, A.; Kreutz, M.; Knuchel, R.; Krause, S.W.; Andreesen, R. Cytokine repertoire during maturation of monocytes to macrophages within spheroids of malignant and non-malignant urothelial cell lines. Int. J. Cancer 1998, 78, 648–653. [Google Scholar] [CrossRef]
- Gottfried, E.; Faust, S.; Fritsche, J.; Kunz-Schughart, L.A.; Andreesen, R.; Miyake, K.; Kreutz, M. Identification of genes expressed in tumor-associated macrophages. Immunobiology 2003, 207, 351–359. [Google Scholar] [CrossRef] [PubMed]
- Ward, J.P.; King, J.R. Mathematical modelling of drug transport in tumour multicell spheroids and monolayer cultures. Math. Biosci. 2003, 181, 177–207. [Google Scholar] [CrossRef]
- Wenning, L.A.; Murphy, R.M. Coupled cellular trafficking and diffusional limitations in delivery of immunotoxins to multicell tumor spheroids. Biotechnol. Bioeng. 1999, 62, 562–575. [Google Scholar] [CrossRef]
- Abu-Absi, S.F.; Friend, J.R.; Hansen, L.K.; Hu, W.S. Structural polarity and functional bile canaliculi in rat hepatocyte spheroids. Exp. Cell. Res. 2002, 274, 56–67. [Google Scholar] [CrossRef] [PubMed]
- Tong, J.Z.; Sarrazin, S.; Cassio, D.; Gauthier, F.; Alvarez, F. Application of spheroid culture to human hepatocytes and maintenance of their differentiation. Biol. Cell. 1994, 81, 77–81. [Google Scholar] [CrossRef]
- Anderer, U.; Libera, J. In vitro engineering of human autogenous cartilage. J. Bone Miner. Res. 2002, 17, 1420–1429. [Google Scholar] [CrossRef] [PubMed]
- Jedeszko, C.; Sameni, M.; Olive, M.B.; Moin, K.; Sloane, B.F. Visualizing protease activity in living cells: From two dimensions to four dimensions. Curr. Protoc. Cell. Biol. 2008. [Google Scholar] [CrossRef]
- Bai, C.; Yang, M.; Fan, Z.; Li, S.; Gao, T.; Fang, Z. Associations of chemo- and radio-resistant phenotypes with the gap junction, adhesion and extracellular matrix in a three-dimensional culture model of soft sarcoma. J. Exp. Clin. Cancer Res. 2015, 34, 58. [Google Scholar] [CrossRef] [PubMed]
- Sachs, N.; Clevers, H. Organoid cultures for the analysis of cancer phenotypes. Curr. Opin. Genet. Dev. 2014, 24, 68–73. [Google Scholar] [CrossRef] [PubMed]
- Halfter, K.; Ditsch, N.; Kolberg, H.C.; Fischer, H.; Hauzenberger, T.; von Koch, F.E.; Bauerfeind, I.; von Minckwitz, G.; Funke, I.; Crispin, A.; et al. Prospective cohort study using the breast cancer spheroid model as a predictor for response to neoadjuvant therapy—The spheroneo study. BMC Cancer 2015, 15, 519. [Google Scholar] [CrossRef] [PubMed]
- Van Oosterwijk, J.G.; Herpers, B.; Meijer, D.; Briaire-de Bruijn, I.H.; Cleton-Jansen, A.M.; Gelderblom, H.; van de Water, B.; Bovee, J.V. Restoration of chemosensitivity for doxorubicin and cisplatin in chondrosarcoma in vitro: Bcl-2 family members cause chemoresistance. Ann. Oncol 2012, 23, 1617–1626. [Google Scholar] [CrossRef] [PubMed]
- Monderer, D.; Luseau, A.; Bellec, A.; David, E.; Ponsolle, S.; Saiagh, S.; Bercegeay, S.; Piloquet, P.; Denis, M.G.; Lode, L.; et al. New chondrosarcoma cell lines and mouse models to study the link between chondrogenesis and chemoresistance. Lab. Invest. 2013, 93, 1100–1114. [Google Scholar] [CrossRef] [PubMed]
- Hamdi, D.H.; Barbieri, S.; Chevalier, F.; Groetz, J.E.; Legendre, F.; Demoor, M.; Galera, P.; Lefaix, J.L.; Saintigny, Y. In vitro engineering of human 3D chondrosarcoma: A preclinical model relevant for investigations of radiation quality impact. BMC Cancer 2015, 15, 579. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pauli, C.; Hopkins, B.D.; Prandi, D.; Shaw, R.; Fedrizzi, T.; Sboner, A.; Sailer, V.; Augello, M.; Puca, L.; Rosati, R.; et al. Personalized in vitro and in vivo cancer models to guide precision medicine. Cancer Discov. 2017, 7, 462–477. [Google Scholar] [CrossRef] [PubMed]
- Boehnke, K.; Iversen, P.W.; Schumacher, D.; Lallena, M.J.; Haro, R.; Amat, J.; Haybaeck, J.; Liebs, S.; Lange, M.; Schafer, R.; et al. Assay establishment and validation of a high-throughput screening platform for three-dimensional patient-derived colon cancer organoid cultures. J. Biomol. Screen. 2016, 21, 931–941. [Google Scholar] [CrossRef] [PubMed]
- Breslin, S.; O'Driscoll, L. Three-dimensional cell culture: The missing link in drug discovery. Drug Discov. Today 2013, 18, 240–249. [Google Scholar] [CrossRef] [PubMed]
- Ricci, C.; Moroni, L.; Danti, S. Cancer tissue engineering—New perspectives in understanding the biology of solid tumours—A critical review. OA Tissue Eng. 2013, 1, 4. [Google Scholar] [CrossRef]
- Tibbitt, M.W.; Anseth, K.S. Hydrogels as extracellular matrix mimics for 3D cell culture. Biotechnol. Bioeng. 2009, 103, 655–663. [Google Scholar] [CrossRef] [PubMed]
- Fischbach, C.; Chen, R.; Matsumoto, T.; Schmelzle, T.; Brugge, J.S.; Polverini, P.J.; Mooney, D.J. Engineering tumors with 3D scaffolds. Nat. Methods 2007, 4, 855–860. [Google Scholar] [CrossRef] [PubMed]

| Models | General Properties/Advantages | Cost and Reproducibility | Disadvantages | References |
|---|---|---|---|---|
| 2D cell cultures Monolayers cultured on polystyrene or other adherent flat surfaces |
|
|
| [108,109] |
| 3D scaffold-based cell cultures Cells seeded in structures of different materials: hydrogels, 3D life biomimetic, 3D Insert™ scaffolds (synthetic bone derivatives), Alvetex® |
|
|
| [109,110,111] |
| 3D nonscaffold-based cell cultures Spheroids from single or multiple cell types; Low-adhesion plates; Micropatterned surfaces (Scivax NanoCulture® multiwell plates); Hanging drop plates (Perfecta3D®, Gravity-PLUS™ 3D) |
|
|
| [108,109] |
| Animal models Patient-derivative xenograph (PDX) |
|
|
| [47,70] |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Colella, G.; Fazioli, F.; Gallo, M.; De Chiara, A.; Apice, G.; Ruosi, C.; Cimmino, A.; De Nigris, F. Sarcoma Spheroids and Organoids—Promising Tools in the Era of Personalized Medicine. Int. J. Mol. Sci. 2018, 19, 615. https://doi.org/10.3390/ijms19020615
Colella G, Fazioli F, Gallo M, De Chiara A, Apice G, Ruosi C, Cimmino A, De Nigris F. Sarcoma Spheroids and Organoids—Promising Tools in the Era of Personalized Medicine. International Journal of Molecular Sciences. 2018; 19(2):615. https://doi.org/10.3390/ijms19020615
Chicago/Turabian StyleColella, Gianluca, Flavio Fazioli, Michele Gallo, Annarosaria De Chiara, Gaetano Apice, Carlo Ruosi, Amelia Cimmino, and Filomena De Nigris. 2018. "Sarcoma Spheroids and Organoids—Promising Tools in the Era of Personalized Medicine" International Journal of Molecular Sciences 19, no. 2: 615. https://doi.org/10.3390/ijms19020615
APA StyleColella, G., Fazioli, F., Gallo, M., De Chiara, A., Apice, G., Ruosi, C., Cimmino, A., & De Nigris, F. (2018). Sarcoma Spheroids and Organoids—Promising Tools in the Era of Personalized Medicine. International Journal of Molecular Sciences, 19(2), 615. https://doi.org/10.3390/ijms19020615