Cell-Penetrating Function of the Poly(ADP-Ribose) (PAR)-Binding Motif Derived from the PAR-Dependent E3 Ubiquitin Ligase Iduna
Abstract
:1. Introduction
2. Results
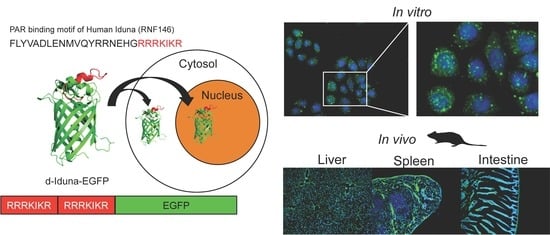
2.1. CPP Candidate Sequence in Iduna Protein
2.2. Protein Delivery in Jurkat T Cells by the Iduna-Derived Sequence
2.3. Intracellular Protein Delivery Mechanism of the Iduna-Derived Sequence
2.4. In Vitro Localization of the Iduna-Derived Sequence
2.5. In Vivo Tissue and Cellular Localization of d-Iduna-EGFP in Mice
3. Discussion
4. Materials and Methods
4.1. Recombinant Protein Purification
4.2. 3D Structure Modeling
4.3. Cell Lines and Cell Culture
4.4. In Vitro Delivery Efficiency
4.5. Mechanism of Internalization
4.6. Intracellular Localization
4.7. Mice
4.8. Cytotoxicity Assay
4.9. Statistics
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Koren, E.; Torchilin, V.P. Cell-penetrating peptides: Breaking through to the other side. Trends Mol. Med. 2012, 18, 385–393. [Google Scholar] [CrossRef] [PubMed]
- Snyder, E.L.; Dowdy, S.F. Cell penetrating peptides in drug delivery. Pharm. Res. 2004, 21, 389–393. [Google Scholar] [CrossRef] [PubMed]
- Zorko, M.; Langel, U. Cell-penetrating peptides: Mechanism and kinetics of cargo delivery. Adv. Drug Deliv. Rev. 2005, 57, 529–545. [Google Scholar] [CrossRef] [PubMed]
- Frankel, A.D.; Pabo, C.O. Cellular uptake of the tat protein from human immunodeficiency virus. Cell 1988, 55, 1189–1193. [Google Scholar] [CrossRef]
- Green, M.; Loewenstein, P.M. Autonomous functional domains of chemically synthesized human immunodeficiency virus tat trans-activator protein. Cell 1988, 55, 1179–1188. [Google Scholar] [CrossRef]
- Schwarze, S.R.; Ho, A.; Vocero-Akbani, A.; Dowdy, S.F. In vivo protein transduction: Delivery of a biologically active protein into the mouse. Science 1999, 285, 1569–1572. [Google Scholar] [CrossRef] [PubMed]
- Derossi, D.; Calvet, S.; Trembleau, A.; Brunissen, A.; Chassaing, G.; Prochiantz, A. Cell internalization of the third helix of the Antennapedia homeodomain is receptor-independent. J. Biol. Chem. 1996, 271, 18188–18193. [Google Scholar] [CrossRef] [PubMed]
- Derossi, D.; Joliot, A.H.; Chassaing, G.; Prochiantz, A. The third helix of the Antennapedia homeodomain translocates through biological membranes. J. Biol. Chem. 1994, 269, 10444–10450. [Google Scholar] [PubMed]
- Elliott, G.; O’Hare, P. Intercellular trafficking and protein delivery by a herpesvirus structural protein. Cell 1997, 88, 223–233. [Google Scholar] [CrossRef]
- Pooga, M.; Hallbrink, M.; Zorko, M.; Langel, U. Cell penetration by transportan. FASEB J. 1998, 12, 67–77. [Google Scholar] [CrossRef] [PubMed]
- Futaki, S.; Suzuki, T.; Ohashi, W.; Yagami, T.; Tanaka, S.; Ueda, K.; Sugiura, Y. Arginine-rich peptides. An abundant source of membrane-permeable peptides having potential as carriers for intracellular protein delivery. J. Biol. Chem. 2001, 276, 5836–5840. [Google Scholar] [CrossRef] [PubMed]
- Nakase, I.; Niwa, M.; Takeuchi, T.; Sonomura, K.; Kawabata, N.; Koike, Y.; Takehashi, M.; Tanaka, S.; Ueda, K.; Simpson, J.C.; et al. Cellular uptake of arginine-rich peptides: Roles for macropinocytosis and actin rearrangement. Mol. Ther. 2004, 10, 1011–1022. [Google Scholar] [CrossRef] [PubMed]
- Choi, J.M.; Ahn, M.H.; Chae, W.J.; Jung, Y.G.; Park, J.C.; Song, H.M.; Kim, Y.E.; Shin, J.A.; Park, C.S.; Park, J.W.; et al. Intranasal delivery of the cytoplasmic domain of CTLA-4 using a novel protein transduction domain prevents allergic inflammation. Nat. Med. 2006, 12, 574–579. [Google Scholar] [CrossRef] [PubMed]
- Choi, J.M.; Kim, S.H.; Shin, J.H.; Gibson, T.; Yoon, B.S.; Lee, D.H.; Lee, S.K.; Bothwell, A.L.; Lim, J.S.; Lee, S.K. Transduction of the cytoplasmic domain of CTLA-4 inhibits TcR-specific activation signals and prevents collagen-induced arthritis. Proc. Natl. Acad. Sci. USA 2008, 105, 19875–19880. [Google Scholar] [CrossRef] [PubMed]
- Choi, J.M.; Sohn, J.H.; Park, T.Y.; Park, J.W.; Lee, S.K. Cell permeable NFAT inhibitory peptide Sim-2-VIVIT inhibits T-cell activation and alleviates allergic airway inflammation and hyper-responsiveness. Immunol. Lett. 2012, 143, 170–176. [Google Scholar] [CrossRef] [PubMed]
- Fang, S.L.; Fan, T.C.; Fu, H.W.; Chen, C.J.; Hwang, C.S.; Hung, T.J.; Lin, L.Y.; Chang, M.D. A novel cell-penetrating peptide derived from human eosinophil cationic protein. PLoS ONE 2013, 8, e57318. [Google Scholar] [CrossRef] [PubMed]
- Harreither, E.; Rydberg, H.A.; Amand, H.L.; Jadhav, V.; Fliedl, L.; Benda, C.; Esteban, M.A.; Pei, D.; Borth, N.; Grillari-Voglauer, R.; et al. Characterization of a novel cell penetrating peptide derived from human Oct4. Cell Regen. 2014, 3, 2. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lim, S.; Kim, W.J.; Kim, Y.H.; Choi, J.M. Identification of a novel cell-penetrating peptide from human phosphatidate phosphatase LPIN3. Mol. Cells 2012, 34, 577–582. [Google Scholar] [CrossRef] [PubMed]
- Koo, J.H.; Yoon, H.; Kim, W.J.; Lim, S.; Park, H.J.; Choi, J.M. Cell membrane penetrating function of the nuclear localization sequence in human cytokine IL-1alpha. Mol. Biol. Rep. 2014, 41, 8117–8126. [Google Scholar] [CrossRef] [PubMed]
- Lim, S.; Kim, W.J.; Kim, Y.H.; Lee, S.; Koo, J.H.; Lee, J.A.; Yoon, H.; Kim, D.H.; Park, H.J.; Kim, H.M.; et al. dNP2 is a blood-brain barrier-permeable peptide enabling ctCTLA-4 protein delivery to ameliorate experimental autoimmune encephalomyelitis. Nat. Commun. 2015, 6, 8244. [Google Scholar] [CrossRef] [PubMed]
- Kim, W.J.; Koo, J.H.; Cho, H.J.; Lee, J.U.; Kim, J.Y.; Lee, H.G.; Lee, S.; Kim, J.H.; Oh, M.S.; Suh, M.; et al. Protein tyrosine phosphatase conjugated with a novel transdermal delivery peptide, astrotactin 1-derived peptide recombinant protein tyrosine phosphatase (AP-rPTP), alleviates both atopic dermatitis-like and psoriasis-like dermatitis. J. Allergy Clin. Immunol. 2018, 141, 137–151. [Google Scholar] [CrossRef] [PubMed]
- Gibson, B.A.; Kraus, W.L. New insights into the molecular and cellular functions of poly(ADP-ribose) and PARPs. Nat. Rev. Mol. Cell Biol. 2012, 13, 411–424. [Google Scholar] [CrossRef] [PubMed]
- Herceg, Z.; Wang, Z.Q. Functions of poly(ADP-ribose) polymerase (PARP) in DNA repair, genomic integrity and cell death. Mutat. Res. 2001, 477, 97–110. [Google Scholar] [CrossRef]
- Wei, H.; Yu, X. Functions of PARylation in DNA Damage Repair Pathways. Genom. Proteom. Bioinformat. 2016, 14, 131–139. [Google Scholar] [CrossRef] [PubMed]
- Kang, H.C.; Lee, Y.I.; Shin, J.H.; Andrabi, S.A.; Chi, Z.; Gagne, J.P.; Lee, Y.; Ko, H.S.; Lee, B.D.; Poirier, G.G.; et al. Iduna is a poly(ADP-ribose) (PAR)-dependent E3 ubiquitin ligase that regulates DNA damage. Proc. Natl. Acad. Sci. USA 2011, 108, 14103–14108. [Google Scholar] [CrossRef] [PubMed]
- Andrabi, S.A.; Kang, H.C.; Haince, J.F.; Lee, Y.I.; Zhang, J.; Chi, Z.; West, A.B.; Koehler, R.C.; Poirier, G.G.; Dawson, T.M.; et al. Iduna protects the brain from glutamate excitotoxicity and stroke by interfering with poly(ADP-ribose) polymer-induced cell death. Nat. Med. 2011, 17, 692–699. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Z.D.; Chan, C.H.; Xiao, Z.C.; Tan, E.K. Ring finger protein 146/Iduna is a poly(ADP-ribose) polymer binding and PARsylation dependent E3 ubiquitin ligase. Cell Adhes. Migr. 2011, 5, 463–471. [Google Scholar] [CrossRef] [PubMed]
- Gautam, A.; Chaudhary, K.; Kumar, R.; Sharma, A.; Kapoor, P.; Tyagi, A.; Open Source Drug Discovery Consortium; Raghava, G.P. In silico approaches for designing highly effective cell penetrating peptides. J. Transl. Med. 2013, 11, 74. [Google Scholar] [CrossRef] [PubMed]
- Goncalves, E.; Kitas, E.; Seelig, J. Binding of oligoarginine to membrane lipids and heparan sulfate: Structural and thermodynamic characterization of a cell-penetrating peptide. Biochemistry 2005, 44, 2692–2702. [Google Scholar] [CrossRef] [PubMed]
- Richard, J.P.; Melikov, K.; Brooks, H.; Prevot, P.; Lebleu, B.; Chernomordik, L.V. Cellular uptake of unconjugated TAT peptide involves clathrin-dependent endocytosis and heparan sulfate receptors. J. Biol. Chem. 2005, 280, 15300–15306. [Google Scholar] [CrossRef] [PubMed]
- Tyagi, M.; Rusnati, M.; Presta, M.; Giacca, M. Internalization of HIV-1 tat requires cell surface heparan sulfate proteoglycans. J. Biol. Chem. 2001, 276, 3254–3261. [Google Scholar] [CrossRef] [PubMed]
- Jiao, C.Y.; Delaroche, D.; Burlina, F.; Alves, I.D.; Chassaing, G.; Sagan, S. Translocation and endocytosis for cell-penetrating peptide internalization. J. Biol. Chem. 2009, 284, 33957–33965. [Google Scholar] [CrossRef] [PubMed]
- El-Sayed, A.; Futaki, S.; Harashima, H. Delivery of macromolecules using arginine-rich cell-penetrating peptides: Ways to overcome endosomal entrapment. AAPS J. 2009, 11, 13–22. [Google Scholar] [CrossRef] [PubMed]
- Madani, F.; Lindberg, S.; Langel, U.; Futaki, S.; Graslund, A. Mechanisms of cellular uptake of cell-penetrating peptides. J. Biophys. 2011, 2011, 414729. [Google Scholar] [CrossRef] [PubMed]
- Guidotti, G.; Brambilla, L.; Rossi, D. Cell-Penetrating Peptides: From Basic Research to Clinics. Trends Pharmacol. Sci. 2017, 38, 406–424. [Google Scholar] [CrossRef] [PubMed]
- Trehin, R.; Merkle, H.P. Chances and pitfalls of cell penetrating peptides for cellular drug delivery. Eur. J. Pharm. Biopharm. 2004, 58, 209–223. [Google Scholar] [CrossRef] [PubMed]
- Choi, J.M.; Shin, J.H.; Sohn, M.H.; Harding, M.J.; Park, J.H.; Tobiasova, Z.; Kim, D.Y.; Maher, S.E.; Chae, W.J.; Park, S.H.; et al. Cell-permeable Foxp3 protein alleviates autoimmune disease associated with inflammatory bowel disease and allergic airway inflammation. Proc. Natl. Acad. Sci. USA 2010, 107, 18575–18580. [Google Scholar] [CrossRef] [PubMed]
- Leppard, J.B.; Dong, Z.; Mackey, Z.B.; Tomkinson, A.E. Physical and functional interaction between DNA ligase IIIalpha and poly(ADP-Ribose) polymerase 1 in DNA single-strand break repair. Mol. Cell Biol. 2003, 23, 5919–5927. [Google Scholar] [CrossRef] [PubMed]
- Fisher, A.E.; Hochegger, H.; Takeda, S.; Caldecott, K.W. Poly(ADP-ribose) polymerase 1 accelerates single-strand break repair in concert with poly(ADP-ribose) glycohydrolase. Mol. Cell Biol. 2007, 27, 5597–5605. [Google Scholar] [CrossRef] [PubMed]










© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Koo, J.-H.; Yoon, H.; Kim, W.-J.; Cha, D.; Choi, J.-M. Cell-Penetrating Function of the Poly(ADP-Ribose) (PAR)-Binding Motif Derived from the PAR-Dependent E3 Ubiquitin Ligase Iduna. Int. J. Mol. Sci. 2018, 19, 779. https://doi.org/10.3390/ijms19030779
Koo J-H, Yoon H, Kim W-J, Cha D, Choi J-M. Cell-Penetrating Function of the Poly(ADP-Ribose) (PAR)-Binding Motif Derived from the PAR-Dependent E3 Ubiquitin Ligase Iduna. International Journal of Molecular Sciences. 2018; 19(3):779. https://doi.org/10.3390/ijms19030779
Chicago/Turabian StyleKoo, Ja-Hyun, Heeseok Yoon, Won-Ju Kim, Donghun Cha, and Je-Min Choi. 2018. "Cell-Penetrating Function of the Poly(ADP-Ribose) (PAR)-Binding Motif Derived from the PAR-Dependent E3 Ubiquitin Ligase Iduna" International Journal of Molecular Sciences 19, no. 3: 779. https://doi.org/10.3390/ijms19030779
APA StyleKoo, J. -H., Yoon, H., Kim, W. -J., Cha, D., & Choi, J. -M. (2018). Cell-Penetrating Function of the Poly(ADP-Ribose) (PAR)-Binding Motif Derived from the PAR-Dependent E3 Ubiquitin Ligase Iduna. International Journal of Molecular Sciences, 19(3), 779. https://doi.org/10.3390/ijms19030779