Non-Coding RNA in the Pathogenesis, Progression and Treatment of Hypertension
Abstract
:1. Introduction
2. Discovery and Application of Non-Coding RNAs
3. Recent Progress of miRNAs in Hypertension
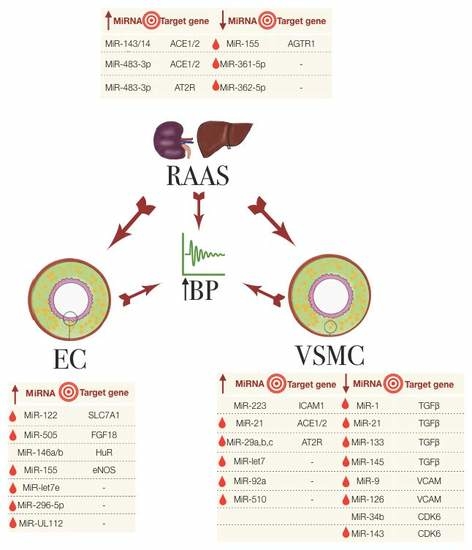
3.1. miRNAs in the Regulation of the Renin-Angiotensin Aldosterone System
3.2. miRNAs in Endothelial Dysfunction
3.3. miRNAs Involved in VSMCs and Other Cells in Hypertension
4. Recent Progress of lncRNAs in Hypertension
5. Detection of Non-Coding RNAs
6. Non-Coding RNAs in the Treatment of Hypertension
7. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Mills, K.T.; Bundy, J.D.; Kelly, T.N.; Reed, J.E.; Kearney, P.M.; Reynolds, K.; Chen, J.; He, J. Global Disparities of Hypertension Prevalence and Control: A Systematic Analysis of Population-Based Studies From 90 Countries. Circulation 2016, 134, 441–450. [Google Scholar] [CrossRef] [PubMed]
- Rosendorff, C.; Lackland, D.T.; Allison, M.; Aronow, W.S.; Black, H.R.; Blumenthal, R.S.; Cannon, C.P.; de Lemos, J.A.; Elliott, W.J.; Findeiss, L.; et al. Treatment of Hypertension in Patients With Coronary Artery Disease. A Scientific Statement From the American Heart Association, American College of Cardiology, and American Society of Hypertension. Circulation 2015, 131, e435–e470. [Google Scholar] [CrossRef] [PubMed]
- Whelton, P.K.; Carey, R.M.; Aronow, W.S.; Casey, D.E.; Collins, K.J.; Dennison Himmelfarb, C.; DePalma, S.M.; Gidding, S.; Jamerson, K.A.; Jones, D.W.; et al. 2017 ACC/AHA/AAPA/ABC/ACPM/AGS/APhA/ASH/ASPC/NMA/PCNA Guideline for the Prevention, Detection, Evaluation, and Management of High Blood Pressure in Adults. A Report of the American College of Cardiology/American Heart Association Task Force on Clinical Practice Guidelines. Hypertension 2017. [Google Scholar] [CrossRef]
- Carretero, O.A.; Oparil, S. Essential Hypertension. Part I: Definition and Etiology. Circulation 2000, 101, 329–335. [Google Scholar] [CrossRef] [PubMed]
- Poulter, N.R.; Prabhakaran, D.; Caulfield, M. Hypertension. Lancet 2015, 386, 801–812. [Google Scholar] [CrossRef]
- Derhaschnig, U.; Testori, C.; Riedmueller, E.; Aschauer, S.; Wolzt, M.; Jilma, B. Hypertensive emergencies are associated with elevated markers of inflammation, coagulation, platelet activation and fibrinolysis. J. Hum. Hypertens. 2012, 27, 368–373. [Google Scholar] [CrossRef] [PubMed]
- Schlaich, M.P.; Lambert, E.; Kaye, D.M.; Krozowski, Z.; Campbell, D.J.; Lambert, G.; Hastings, J.; Aggarwal, A.; Esler, M.D. Sympathetic augmentation in hypertension: Role of nerve firing, norepinephrine reuptake, and Angiotensin neuromodulation. Hypertension 2004, 43, 169–175. [Google Scholar] [CrossRef] [PubMed]
- Cadwgan, T.M.; Benjamin, N. Evidence for altered platelet nitric oxide synthesis in essential hypertension. J. Hypertens. 1993, 11, 417–420. [Google Scholar] [CrossRef] [PubMed]
- Brunini, T.; Moss, M.; Siqueira, M.; Meirelles, L.; Rozentul, A.; Mann, G.; Ellory, J.; Soares de Moura, R.; Mendes-Ribeiro, A. Inhibition of l-arginine transport in platelets by asymmetric dimethylarginine and N-monomethyl-l-arginine: Effects of arterial hypertension. Clin. Exp. Pharmacol. Physiol. 2004, 31, 738–740. [Google Scholar] [CrossRef] [PubMed]
- Perticone, F.; Sciacqua, A.; Maio, R.; Perticone, M.; Maas, R.; Boger, R.H.; Tripepi, G.; Sesti, G.; Zoccali, C. Asymmetric dimethylarginine, l-arginine, and endothelial dysfunction in essential hypertension. J. Am. Coll. Cardiol. 2005, 46, 518–523. [Google Scholar] [CrossRef] [PubMed]
- Bavishi, C.; Bangalore, S.; Messerli, F.H. Outcomes of Intensive Blood Pressure Lowering in Older Hypertensive Patients. J. Am. Coll. Cardiol. 2017, 69, 486–493. [Google Scholar] [CrossRef] [PubMed]
- Patel, R.S.; Masi, S.; Taddei, S. Understanding the role of genetics in hypertension. Eur. Heart J. 2017, 38, 2309–2312. [Google Scholar] [CrossRef] [PubMed]
- Kellis, M.; Wold, B.; Snyder, M.P.; Bernstein, B.E.; Kundaje, A.; Marinov, G.K.; Ward, L.D.; Birney, E.; Crawford, G.E.; Dekker, J.; et al. Defining functional DNA elements in the human genome. Proc. Natl. Acad. Sci. USA 2014, 111, 6131–6138. [Google Scholar] [CrossRef] [PubMed]
- Murakami, K. Non-coding RNAs and hypertension-unveiling unexpected mechanisms of hypertension by the dark matter of the genome. Curr. Hypertens. Rev. 2015, 11, 80–90. [Google Scholar] [CrossRef] [PubMed]
- Batkai, S.; Thum, T. MicroRNAs in hypertension: Mechanisms and therapeutic targets. Curr. Hypertens. Rep. 2012, 14, 79–87. [Google Scholar] [CrossRef] [PubMed]
- Palazzo, A.F.; Lee, E.S. Non-coding RNA: What is functional and what is junk? Front. Genet. 2015, 6, 2. [Google Scholar] [CrossRef] [PubMed]
- Cech, T.R.; Steitz, J.A. The noncoding RNA revolution-trashing old rules to forge new ones. Cell 2014, 157, 77–94. [Google Scholar] [CrossRef] [PubMed]
- Andersen, J.; Delihas, N.; Ikenaka, K.; Green, P.J.; Pines, O.; Ilercil, O.; Inouye, M. The isolation and characterization of RNA coded by the micF gene in Escherichia coli. Nucleic Acids Res. 1987, 15, 2089–2101. [Google Scholar] [CrossRef] [PubMed]
- Andersen, J.; Forst, S.A.; Zhao, K.; Inouye, M.; Delihas, N. The function of micF RNA. micF RNA is a major factor in the thermal regulation of OmpF protein in Escherichia coli. J. Biol. Chem. 1989, 264, 17961–17970. [Google Scholar] [PubMed]
- Mizuno, T.; Chou, M.Y.; Inouye, M. A unique mechanism regulating gene expression: Translational inhibition by a complementary RNA transcript (micRNA). Proc. Natl. Acad. Sci. USA 1984, 81, 1966–1970. [Google Scholar] [CrossRef] [PubMed]
- Lee, R.C.; Feinbaum, R.L.; Ambros, V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 1993, 75, 843–854. [Google Scholar] [CrossRef]
- Brockdorff, N.; Ashworth, A.; Kay, G.F.; McCabe, V.M.; Norris, D.P.; Cooper, P.J.; Swift, S.; Rastan, S. The product of the mouse Xist gene is a 15 kb inactive X-specific transcript containing no conserved ORF and located in the nucleus. Cell 1992, 71, 515–526. [Google Scholar] [CrossRef]
- Pasquinelli, A.E.; Reinhart, B.J.; Slack, F.; Martindale, M.Q.; Kuroda, M.I.; Maller, B.; Hayward, D.C.; Ball, E.E.; Degnan, B.; Müller, P.; et al. Conservation of the sequence and temporal expression of let-7 heterochronic regulatory RNA. Nature 2000, 408, 86–89. [Google Scholar] [PubMed]
- Reinhart, B.J.; Slack, F.J.; Basson, M.; Pasquinelli, A.E.; Bettinger, J.C.; Rougvie, A.E.; Horvitz, H.R.; Ruvkun, G. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature 2000, 403, 901–906. [Google Scholar] [CrossRef] [PubMed]
- Calin, G.A.; Dumitru, C.D.; Shimizu, M.; Bichi, R.; Zupo, S.; Noch, E.; Aldler, H.; Rattan, S.; Keating, M.; Rai, K.; et al. Frequent deletions and down-regulation of micro-RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc. Natl. Acad. Sci. USA 2002, 99, 15524–15529. [Google Scholar] [CrossRef] [PubMed]
- Eis, P.S.; Tam, W.; Sun, L.; Chadburn, A.; Li, Z.; Gomez, M.F.; Lund, E.; Dahlberg, J.E. Accumulation of miR-155 and BIC RNA in human B cell lymphomas. Proc. Natl. Acad. Sci. USA 2005, 102, 3627–3632. [Google Scholar] [CrossRef] [PubMed]
- He, L.; Thomson, J.M.; Hemann, M.T.; Hernando-Monge, E.; Mu, D.; Goodson, S.; Powers, S.; Cordon-Cardo, C.; Lowe, S.W.; Hannon, G.J.; et al. A microRNA polycistron as a potential human oncogene. Nature 2005, 435, 828–833. [Google Scholar] [CrossRef] [PubMed]
- Costinean, S.; Zanesi, N.; Pekarsky, Y.; Tili, E.; Volinia, S.; Heerema, N.; Croce, C.M. Pre-B cell proliferation and lymphoblastic leukemia/high-grade lymphoma in Eμ-miR155 transgenic mice. Proc. Natl. Acad. Sci. USA 2006, 103, 7024–7029. [Google Scholar] [CrossRef] [PubMed]
- Daka, A.; Peer, D. RNAi-based nanomedicines for targeted personalized therapy. Adv. Drug Deliv. Rev. 2012, 64, 1508–1521. [Google Scholar] [CrossRef] [PubMed]
- Lam, J.K.W.; Chow, M.Y.T.; Zhang, Y.; Leung, S.W.S. siRNA Versus miRNA as Therapeutics for Gene Silencing. Mol. Ther. Nucleic Acids 2015, 4, e252. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chakraborty, C.; Sharma, A.R.; Sharma, G.; Doss, C.G.P.; Lee, S.-S. Therapeutic miRNA and siRNA: Moving from Bench to Clinic as Next Generation Medicine. Mol. Ther. Nucleic Acids 2017, 8, 132–143. [Google Scholar] [CrossRef] [PubMed]
- Gebert, L.F.; Rebhan, M.A.; Crivelli, S.E.; Denzler, R.; Stoffel, M.; Hall, J. Miravirsen (SPC3649) can inhibit the biogenesis of miR-122. Nucleic Acids Res. 2014, 42, 609–621. [Google Scholar] [CrossRef] [PubMed]
- Janssen, H.L.; Reesink, H.W.; Lawitz, E.J.; Zeuzem, S.; Rodriguez-Torres, M.; Patel, K.; van der Meer, A.J.; Patick, A.K.; Chen, A.; Zhou, Y.; et al. Treatment of HCV infection by targeting microRNA. N. Engl. J. Med. 2013, 368, 1685–1694. [Google Scholar] [CrossRef] [PubMed]
- Hergenreider, E.; Heydt, S.; Tréguer, K.; Boettger, T.; Horrevoets, A.J.G.; Zeiher, A.M.; Scheffer, M.P.; Frangakis, A.S.; Yin, X.; Mayr, M.; et al. Atheroprotective communication between endothelial cells and smooth muscle cells through miRNAs. Nat. Cell Biol. 2012, 14, 249–256. [Google Scholar] [CrossRef] [PubMed]
- Vickers, K.C.; Palmisano, B.T.; Shoucri, B.M.; Shamburek, R.D.; Remaley, A.T. MicroRNAs are transported in plasma and delivered to recipient cells by high-density lipoproteins. Nat. Cell Biol. 2011, 13, 423–433. [Google Scholar] [CrossRef] [PubMed]
- Valadi, H.; Ekström, K.; Bossios, A.; Sjöstrand, M.; Lee, J.J.; Lötvall, J.O. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat. Cell Biol. 2007, 9, 654–659. [Google Scholar] [CrossRef] [PubMed]
- Arroyo, J.D.; Chevillet, J.R.; Kroh, E.M.; Ruf, I.K.; Pritchard, C.C.; Gibson, D.F.; Mitchell, P.S.; Bennett, C.F.; Pogosova-Agadjanyan, E.L.; Stirewalt, D.L.; et al. Argonaute2 complexes carry a population of circulating microRNAs independent of vesicles in human plasma. Proc. Natl. Acad. Sci. USA 2011, 108, 5003–5008. [Google Scholar] [CrossRef] [PubMed]
- Coffman, T.M.; Crowley, S.D. Kidney in Hypertension: Guyton Redux. Hypertension 2008, 51, 811–816. [Google Scholar] [CrossRef] [PubMed]
- Ceolotto, G.; Papparella, I.; Bortoluzzi, A.; Strapazzon, G.; Ragazzo, F.; Bratti, P.; Fabricio, A.S.C.; Squarcina, E.; Gion, M.; Palatini, P.; et al. Interplay Between miR-155, AT1R A1166C Polymorphism, and AT1R Expression in Young Untreated Hypertensives. Am. J. Hypertens. 2011, 24, 241–246. [Google Scholar] [CrossRef] [PubMed]
- Sethupathy, P.; Borel, C.; Gagnebin, M.; Grant, G.R.; Deutsch, S.; Elton, T.S.; Hatzigeorgiou, A.G.; Antonarakis, S.E. Human microRNA-155 on chromosome 21 differentially interacts with its polymorphic target in the AGTR1 3′ untranslated region: A mechanism for functional single-nucleotide polymorphisms related to phenotypes. Am. J. Hum. Genet. 2007, 81, 405–413. [Google Scholar] [CrossRef] [PubMed]
- Nossent, A.Y.; Hansen, J.L.; Doggen, C.; Quax, P.H.A.; Sheikh, S.P.; Rosendaal, F.R. SNPs in MicroRNA Binding Sites in 3′-UTRs of RAAS Genes Influence Arterial Blood Pressure and Risk of Myocardial Infarction. Am. J. Hypertens. 2011, 24, 999–1006. [Google Scholar] [CrossRef] [PubMed]
- Kemp, J.R.; Unal, H.; Desnoyer, R.; Yue, H.; Bhatnagar, A.; Karnik, S.S. Angiotensin II-regulated microRNA 483-3p directly targets multiple components of the renin-angiotensin system. J. Mol. Cell. Cardiol. 2014, 75, 25–39. [Google Scholar] [CrossRef] [PubMed]
- Kohlstedt, K.; Trouvain, C.; Boettger, T.; Shi, L.; Fisslthaler, B.; Fleming, I. AMP-activated protein kinase regulates endothelial cell angiotensin-converting enzyme expression via p53 and the post-transcriptional regulation of microRNA-143/145. Circ. Res. 2013, 112, 1150–1158. [Google Scholar] [CrossRef] [PubMed]
- Holmberg, J.; Bhattachariya, A.; Alajbegovic, A.; Rippe, C.; Ekman, M.; Dahan, D.; Hien, T.T.; Boettger, T.; Braun, T.; Sward, K.; et al. Loss of Vascular Myogenic Tone in miR-143/145 Knockout Mice Is Associated With Hypertension-Induced Vascular Lesions in Small Mesenteric Arteries. Arterioscler. Thromb. Vasc. Biol. 2018, 38, 414–424. [Google Scholar] [CrossRef] [PubMed]
- Eskildsen, T.; Jeppesen, P.; Schneider, M.; Nossent, A.; Sandberg, M.; Hansen, P.; Jensen, C.; Hansen, M.; Marcussen, N.; Rasmussen, L.; et al. Angiotensin II Regulates microRNA-132/-212 in Hypertensive Rats and Humans. Int. J. Mol. Sci. 2013, 14, 11190–11207. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Romero, D.G.; Plonczynski, M.W.; Carvajal, C.A.; Gomez-Sanchez, E.P.; Gomez-Sanchez, C.E. Microribonucleic Acid-21 Increases Aldosterone Secretion and Proliferation in H295R Human Adrenocortical Cells. Endocrinology 2008, 149, 2477–2483. [Google Scholar] [CrossRef] [PubMed]
- Gildea, J.J.; Carlson, J.M.; Schoeffel, C.D.; Carey, R.M.; Felder, R.A. Urinary exosome miRNome analysis and its applications to salt sensitivity of blood pressure. Clin. Biochem. 2013, 46, 1131–1134. [Google Scholar] [CrossRef] [PubMed]
- Qi, H.; Liu, Z.; Liu, B.; Cao, H.; Sun, W.; Yan, Y.; Zhang, L. micro-RNA screening and prediction model construction for diagnosis of salt-sensitive essential hypertension. Medicine 2017, 96, e6417. [Google Scholar] [CrossRef] [PubMed]
- Marques, F.Z.; Campain, A.E.; Tomaszewski, M.; Zukowska-Szczechowska, E.; Yang, Y.H.; Charchar, F.J.; Morris, B.J. Gene expression profiling reveals renin mRNA overexpression in human hypertensive kidneys and a role for microRNAs. Hypertension 2011, 58, 1093–1098. [Google Scholar] [CrossRef] [PubMed]
- Jackson, K.L.; Marques, F.Z.; Watson, A.M.; Palma-Rigo, K.; Nguyen-Huu, T.P.; Morris, B.J.; Charchar, F.J.; Davern, P.J.; Head, G.A. A novel interaction between sympathetic overactivity and aberrant regulation of renin by miR-181a in BPH/2J genetically hypertensive mice. Hypertension 2013, 62, 775–781. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Kaye, D.M. Mechanistic insights into the link between a polymorphism of the 3′UTR of the SLC7A1 gene and hypertension. Hum. Mutat. 2009, 30, 328–333. [Google Scholar] [CrossRef] [PubMed]
- Yang, Q.; Jia, C.; Wang, P.; Xiong, M.; Cui, J.; Li, L.; Wang, W.; Wu, Q.; Chen, Y.; Zhang, T. MicroRNA-505 identified from patients with essential hypertension impairs endothelial cell migration and tube formation. Int. J. Cardiol. 2014, 177, 925–934. [Google Scholar] [CrossRef] [PubMed]
- Antoine, M.; Wirz, W.; Tag, C.G.; Gressner, A.M.; Wycislo, M.; Müller, R.; Kiefer, P. Fibroblast growth factor 16 and 18 are expressed in human cardiovascular tissues and induce on endothelial cells migration but not proliferation. Biochem. Biophys. Res. Commun. 2006, 346, 224–233. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Zhu, J.; Zhang, W.; Chen, Y.; Zhang, K.; Popescu, L.M.; Ma, X.; Bond Lau, W.; Rong, R.; Yu, X.; et al. Signature microRNA Expression Profile of Essential Hypertension and Its Novel Link to Human Cytomegalovirus Infection. Circulation 2011, 124, 175–184. [Google Scholar] [CrossRef] [PubMed]
- Sun, H.; Zeng, D.; Li, R.; Pang, R.; Yang, H.; Hu, Y.; Zhang, Q.; Jiang, Y.; Huang, L.; Tang, Y.; et al. Essential role of microRNA-155 in regulating endothelium-dependent vasorelaxation by targeting endothelial nitric oxide synthase. Hypertension 2012, 60, 1407–1414. [Google Scholar] [CrossRef] [PubMed]
- Suárez, Y.; Fernández-Hernando, C.; Pober, J.S.; Sessa, W.C. Dicer Dependent MicroRNAs Regulate Gene Expression and Functions in Human Endothelial Cells. Circ. Res. 2007, 100, 1164–1173. [Google Scholar] [CrossRef] [PubMed]
- Dentelli, P.; Rosso, A.; Orso, F.; Olgasi, C.; Taverna, D.; Brizzi, M.F. microRNA-222 controls neovascularization by regulating signal transducer and activator of transcription 5A expression. Arterioscler. Thromb. Vasc. Biol. 2010, 30, 1562–1568. [Google Scholar] [CrossRef] [PubMed]
- Celic, T.; Metzinger-Le Meuth, V.; Six, I.; Massy, Z.A.; Metzinger, L. The mir-221/222 Cluster is a Key Player in Vascular Biology via the Fine-Tuning of Endothelial Cell Physiology. Curr. Vasc. Pharmacol. 2017, 15, 40–46. [Google Scholar] [CrossRef] [PubMed]
- Cheng, H.S.; Sivachandran, N.; Lau, A.; Boudreau, E.; Zhao, J.L.; Baltimore, D.; Delgado-Olguin, P.; Cybulsky, M.I.; Fish, J.E. MicroRNA-146 represses endothelial activation by inhibiting pro-inflammatory pathways. EMBO Mol. Med. 2013, 5, 1017–1034. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Aurora, A.B.; Johnson, B.A.; Qi, X.; McAnally, J.; Hill, J.A.; Richardson, J.A.; Bassel-Duby, R.; Olson, E.N. The Endothelial-Specific MicroRNA miR-126 Governs Vascular Integrity and Angiogenesis. Dev. Cell 2008, 15, 261–271. [Google Scholar] [CrossRef] [PubMed]
- Fish, J.E.; Santoro, M.M.; Morton, S.U.; Yu, S.; Yeh, R.-F.; Wythe, J.D.; Ivey, K.N.; Bruneau, B.G.; Stainier, D.Y.R.; Srivastava, D. miR-126 Regulates Angiogenic Signaling and Vascular Integrity. Dev. Cell 2008, 15, 272–284. [Google Scholar] [CrossRef] [PubMed]
- Togliatto, G.; Trombetta, A.; Dentelli, P.; Gallo, S.; Rosso, A.; Cotogni, P.; Granata, R.; Falcioni, R.; Delale, T.; Ghigo, E.; et al. Unacylated ghrelin induces oxidative stress resistance in a glucose intolerance and peripheral artery disease mouse model by restoring endothelial cell miR-126 expression. Diabetes 2015, 64, 1370–1382. [Google Scholar] [CrossRef] [PubMed]
- Parthenakis, F.; Marketou, M.; Kontaraki, J.; Patrianakos, A.; Nakou, H.; Touloupaki, M.; Vernardos, M.; Kochiadakis, G.; Chlouverakis, G.; Vardas, P. Low Levels of MicroRNA-21 Are a Marker of Reduced Arterial Stiffness in Well-Controlled Hypertension. J. Clin. Hypertens. 2017, 19, 235–240. [Google Scholar] [CrossRef] [PubMed]
- Kontaraki, J.E.; Marketou, M.E.; Zacharis, E.A.; Parthenakis, F.I.; Vardas, P.E. Differential expression of vascular smooth muscle-modulating microRNAs in human peripheral blood mononuclear cells: Novel targets in essential hypertension. J. Hum. Hypertens. 2014, 28, 510–516. [Google Scholar] [CrossRef] [PubMed]
- Zhao, N.; Koenig, S.N.; Trask, A.J.; Lin, C.-H.; Hans, C.P.; Garg, V.; Lilly, B. mir145 Regulates TGFBR2 Expression and Matrix Synthesis in Vascular Smooth Muscle Cells. Circ. Res. 2015, 116, 23–34. [Google Scholar] [CrossRef] [PubMed]
- Kontaraki, J.E.; Marketou, M.E.; Zacharis, E.A.; Parthenakis, F.I.; Vardas, P.E. MicroRNA-9 and microRNA-126 expression levels in patients with essential hypertension: Potential markers of target-organ damage. J. Am. Soc. Hypertens. 2014, 8, 368–375. [Google Scholar] [CrossRef] [PubMed]
- Harris, T.A.; Yamakuchi, M.; Ferlito, M.; Mendell, J.T.; Lowenstein, C.J. MicroRNA-126 regulates endothelial expression of vascular cell adhesion molecule 1. Proc. Natl. Acad. Sci. USA 2008, 105, 1516–1521. [Google Scholar] [CrossRef] [PubMed]
- Tabet, F.; Vickers, K.C.; Cuesta Torres, L.F.; Wiese, C.B.; Shoucri, B.M.; Lambert, G.; Catherinet, C.; Prado-Lourenco, L.; Levin, M.G.; Thacker, S.; et al. HDL-transferred microRNA-223 regulates ICAM-1 expression in endothelial cells. Nat. Commun. 2014, 5, 3292. [Google Scholar] [CrossRef] [PubMed]
- Yang, F.; Li, H.; Du, Y.; Shi, Q.; Zhao, L. Downregulation of microRNA34b is responsible for the elevation of blood pressure in spontaneously hypertensive rats. Mol. Med. Rep. 2017, 15, 1031–1036. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Tang, S.; Huang, C.; Chen, J.; Li, J.; Cai, A.; Feng, Y. Circulating miRNA29 family expression levels in patients with essential hypertension as potential markers for left ventricular hypertrophy. Clin. Exp. Hypertens. 2017, 39, 119–125. [Google Scholar] [CrossRef] [PubMed]
- Krishnan, R.; Mani, P.; Sivakumar, P.; Gopinath, V.; Sekar, D. Expression and methylation of circulating microRNA-510 in essential hypertension. Hypertens. Res. 2017, 40, 361–363. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Huang, C.; Chen, J.; Li, J.; Feng, Y. Plasma expression level of miRNA let-7 is positively correlated with carotid intima-media thickness in patients with essential hypertension. J. Hum. Hypertens. 2017, 31, 843–847. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Tang, S.; Ji-Yan, C.; Huang, C.; Li, J.; Cai, A.; Feng, Y. Circulating miR-92a expression level in patients with essential hypertension: A potential marker of atherosclerosis. J. Hum. Hypertens. 2017, 31, 200–205. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Zhou, Y.; Cao, Z.; Tong, X.; Xie, H.; Luo, T.; Hua, X.; Wang, H. miR-155 functions downstream of angiotensin II receptor subtype 1 and calcineurin to regulate cardiac hypertrophy. Exp. Ther. Med. 2016, 12, 1556–1562. [Google Scholar] [CrossRef] [PubMed]
- Wei, Z.; Biswas, N.; Wang, L.; Courel, M.; Zhang, K.; Soler-Jover, A.; Taupenot, L.; O’Connor, D.T. A common genetic variant in the 3′-UTR of vacuolar H+-ATPase ATP6V0A1 creates a micro-RNA motif to alter chromogranin A processing and hypertension risk. Circ. Cardiovasc. Genet. 2011, 4, 381–389. [Google Scholar] [CrossRef] [PubMed]
- Arora, P.; Wu, C.; Khan, A.M.; Bloch, D.B.; Davis-Dusenbery, B.N.; Ghorbani, A.; Spagnolli, E.; Martinez, A.; Ryan, A.; Tainsh, L.T.; et al. Atrial natriuretic peptide is negatively regulated by microRNA-425. J. Clin. Investig. 2013, 123, 3378–3382. [Google Scholar] [CrossRef] [PubMed]
- Hanin, G.; Shenhar-Tsarfaty, S.; Yayon, N.; Hoe, Y.Y.; Bennett, E.R.; Sklan, E.H.; Rao, D.C.; Rankinen, T.; Bouchard, C.; Geifman-Shochat, S.; et al. Competing targets of microRNA-608 affect anxiety and hypertension. Hum. Mol. Genet. 2014, 23, 4569–4580. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Li, T.; Hao, Y.; Wu, B.; Li, H.; Geng, N.; Sun, Z.; Zheng, L.; Sun, Y. Association of rs2271037 and rs3749585 polymorphisms in CORIN with susceptibility to hypertension in a Chinese Han population: A case-control study. Gene 2018, 651, 79–85. [Google Scholar] [CrossRef] [PubMed]
- Fu, X.; Guo, L.; Jiang, Z.; Zhao, L.; Xu, A. An miR-143 promoter variant associated with essential hypertension. Int. J. Clin. Exp. Med. 2014, 7, 1813–1817. [Google Scholar] [PubMed]
- Boettger, T.; Beetz, N.; Kostin, S.; Schneider, J.; Kruger, M.; Hein, L.; Braun, T. Acquisition of the contractile phenotype by murine arterial smooth muscle cells depends on the Mir143/145 gene cluster. J. Clin. Investig. 2009, 119, 2634–2647. [Google Scholar] [CrossRef] [PubMed]
- Bayoglu, B.; Yuksel, H.; Cakmak, H.A.; Dirican, A.; Cengiz, M. Polymorphisms in the long non-coding RNA CDKN2B-AS1 may contribute to higher systolic blood pressure levels in hypertensive patients. Clin. Biochem. 2016, 49, 821–827. [Google Scholar] [CrossRef] [PubMed]
- Laughlin, M.H.; Newcomer, S.C.; Bender, S.B. Importance of hemodynamic forces as signals for exercise-induced changes in endothelial cell phenotype. J. Appl. Physiol. 2008, 104, 588–600. [Google Scholar] [CrossRef] [PubMed]
- Brandes, R.P. Endothelial Dysfunction and Hypertension. Hypertension 2014, 64, 924–928. [Google Scholar] [CrossRef] [PubMed]
- Chien, S. Mechanotransduction and endothelial cell homeostasis: The wisdom of the cell. Am. J. Physiol. Heart Circ. Physiol. 2007, 292, H1209–H1224. [Google Scholar] [CrossRef] [PubMed]
- Nemecz, M.; Alexandru, N.; Tanko, G.; Georgescu, A. Role of MicroRNA in Endothelial Dysfunction and Hypertension. Curr. Hypertens. Rep. 2016, 18, 87. [Google Scholar] [CrossRef] [PubMed]
- Brozovich, F.V.; Nicholson, C.J.; Degen, C.V.; Gao, Y.Z.; Aggarwal, M.; Morgan, K.G. Mechanisms of Vascular Smooth Muscle Contraction and the Basis for Pharmacologic Treatment of Smooth Muscle Disorders. Pharmacol. Rev. 2016, 68, 476–532. [Google Scholar] [CrossRef] [PubMed]
- Nanoudis, S.; Pikilidou, M.; Yavropoulou, M.; Zebekakis, P. The Role of MicroRNAs in Arterial Stiffness and Arterial Calcification. An Update and Review of the Literature. Front. Genet. 2017, 8, 209. [Google Scholar] [CrossRef] [PubMed]
- Lorenzen, J.M.; Schauerte, C.; Hubner, A.; Kolling, M.; Martino, F.; Scherf, K.; Batkai, S.; Zimmer, K.; Foinquinos, A.; Kaucsar, T.; et al. Osteopontin is indispensible for AP1-mediated angiotensin II-related miR-21 transcription during cardiac fibrosis. Eur. Heart J. 2015, 36, 2184–2196. [Google Scholar] [CrossRef] [PubMed]
- Marquez, R.T.; Bandyopadhyay, S.; Wendlandt, E.B.; Keck, K.; Hoffer, B.A.; Icardi, M.S.; Christensen, R.N.; Schmidt, W.N.; McCaffrey, A.P. Correlation between microRNA expression levels and clinical parameters associated with chronic hepatitis C viral infection in humans. Lab. Investig. 2010, 90, 1727–1736. [Google Scholar] [CrossRef] [PubMed]
- Santovito, D.; Mandolini, C.; Marcantonio, P.; de Nardis, V.; Bucci, M.; Paganelli, C.; Magnacca, F.; Ucchino, S.; Mastroiacovo, D.; Desideri, G.; et al. Overexpression of microRNA-145 in atherosclerotic plaques from hypertensive patients. Expert Opin. Ther. Targets 2013, 17, 217–223. [Google Scholar] [CrossRef] [PubMed]
- Friese, R.S.; Altshuler, A.E.; Zhang, K.; Miramontes-Gonzalez, J.P.; Hightower, C.M.; Jirout, M.L.; Salem, R.M.; Gayen, J.R.; Mahapatra, N.R.; Biswas, N.; et al. MicroRNA-22 and promoter motif polymorphisms at the Chga locus in genetic hypertension: Functional and therapeutic implications for gene expression and the pathogenesis of hypertension. Hum. Mol. Genet. 2013, 22, 3624–3640. [Google Scholar] [CrossRef] [PubMed]
- Bolha, L.; Ravnik-Glavač, M.; Glavač, D. Long Noncoding RNAs as Biomarkers in Cancer. Dis. Markers 2017, 2017, 7243968. [Google Scholar] [CrossRef] [PubMed]
- Yao, Q.P.; Xie, Z.W.; Wang, K.X.; Zhang, P.; Han, Y.; Qi, Y.X.; Jiang, Z.L. Profiles of long noncoding RNAs in hypertensive rats: Long noncoding RNA XR007793 regulates cyclic strain-induced proliferation and migration of vascular smooth muscle cells. J. Hypertens. 2017, 35, 1195–1203. [Google Scholar] [CrossRef] [PubMed]
- Gopalakrishnan, K.; Kumarasamy, S.; Mell, B.; Joe, B. Genome-Wide Identification of Long Noncoding RNAs in Rat Models of Cardiovascular and Renal Disease. Hypertension 2015, 65, 200–210. [Google Scholar] [CrossRef] [PubMed]
- Jin, L.; Lin, X.; Yang, L.; Fan, X.; Wang, W.; Li, S.; Li, J.; Liu, X.; Bao, M.; Cui, X.; et al. AK098656, a Novel Vascular Smooth Muscle Cell–Dominant Long Noncoding RNA, Promotes Hypertension. Hypertension 2018, 71, 262–272. [Google Scholar] [CrossRef] [PubMed]
- Tragante, V.; Barnes, M.R.; Ganesh, S.K.; Lanktree, M.B.; Guo, W.; Franceschini, N.; Smith, E.N.; Johnson, T.; Holmes, M.V.; Padmanabhan, S.; et al. Gene-centric Meta-analysis in 87,736 Individuals of European Ancestry Identifies Multiple Blood-Pressure-Related Loci. Am. J. Hum. Genet. 2014, 94, 349–360. [Google Scholar] [CrossRef] [PubMed]
- Michalik, K.M.; You, X.; Manavski, Y.; Doddaballapur, A.; Zornig, M.; Braun, T.; John, D.; Ponomareva, Y.; Chen, W.; Uchida, S.; et al. Long noncoding RNA MALAT1 regulates endothelial cell function and vessel growth. Circ. Res. 2014, 114, 1389–1397. [Google Scholar] [CrossRef] [PubMed]
- Bell, R.D.; Long, X.; Lin, M.; Bergmann, J.H.; Nanda, V.; Cowan, S.L.; Zhou, Q.; Han, Y.; Spector, D.L.; Zheng, D.; et al. Identification and initial functional characterization of a human vascular cell-enriched long noncoding RNA. Arterioscler. Thromb. Vasc. Biol. 2014, 34, 1249–1259. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Shan, K.; Yao, M.; Yao, J.; Wang, J.; Li, X.; Liu, B.; Zhang, Y.; Ji, Y.; Jiang, Q.; et al. Long Noncoding RNA-GAS5: A Novel Regulator of Hypertension-Induced Vascular Remodeling. Hypertension 2016, 68, 736–748. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Yang, X.; Lin, Y.; Suo, M.; Gong, L.; Chen, J.; Hui, R. Anti-hypertensive effect of Lycium barbarum L. with down-regulated expression of renal endothelial lncRNA sONE in a rat model of salt-sensitive hypertension. Int. J. Clin. Exp. Pathol. 2015, 8, 6981–6987. [Google Scholar] [PubMed]
- Gezer, U.; Ozgur, E.; Cetinkaya, M.; Isin, M.; Dalay, N. Long non-coding RNAs with low expression levels in cells are enriched in secreted exosomes. Cell Biol. Int. 2014, 38, 1076–1079. [Google Scholar] [CrossRef] [PubMed]
- Hewson, C.; Capraro, D.; Burdach, J.; Whitaker, N.; Morris, K.V. Extracellular vesicle associated long non-coding RNAs functionally enhance cell viability. Non-Coding RNA Res. 2016, 1, 3–11. [Google Scholar] [CrossRef] [PubMed]
- Wahlquist, C.; Jeong, D.; Rojas-Muñoz, A.; Kho, C.; Lee, A.; Mitsuyama, S.; van Mil, A.; Jin Park, W.; Sluijter, J.P.G.; Doevendans, P.A.F.; et al. Inhibition of miR-25 improves cardiac contractility in the failing heart. Nature 2014, 508, 531–535. [Google Scholar] [CrossRef] [PubMed]
- Gomes, C.P.C.; Spencer, H.; Ford, K.L.; Michel, L.Y.M.; Baker, A.H.; Emanueli, C.; Balligand, J.-L.; Devaux, Y. The Function and Therapeutic Potential of Long Non-coding RNAs in Cardiovascular Development and Disease. Mol. Ther. Nucleic Acids 2017, 8, 494–507. [Google Scholar] [CrossRef] [PubMed]
- Robb, G.B.; Brown, K.M.; Khurana, J.; Rana, T.M. Specific and potent RNAi in the nucleus of human cells. Nat. Struct. Mol. Biol. 2005, 12, 133–137. [Google Scholar] [CrossRef] [PubMed]
- Bennett, C.F.; Swayze, E.E. RNA targeting therapeutics: Molecular mechanisms of antisense oligonucleotides as a therapeutic platform. Annu. Rev. Pharmacol. Toxicol. 2010, 50, 259–293. [Google Scholar] [CrossRef] [PubMed]
- Arun, G.; Diermeier, S.; Akerman, M.; Chang, K.-C.; Wilkinson, J.E.; Hearn, S.; Kim, Y.; MacLeod, A.R.; Krainer, A.R.; Norton, L.; et al. Differentiation of mammary tumors and reduction in metastasis upon Malat1 lncRNA loss. Genes Dev. 2016, 30, 34–51. [Google Scholar] [CrossRef] [PubMed]
- Fluiter, K.; Mook, O.R.; Vreijling, J.; Langkjaer, N.; Hojland, T.; Wengel, J.; Baas, F. Filling the gap in LNA antisense oligo gapmers: The effects of unlocked nucleic acid (UNA) and 4′-C-hydroxymethyl-DNA modifications on RNase H recruitment and efficacy of an LNA gapmer. Mol. Biosyst. 2009, 5, 838–843. [Google Scholar] [CrossRef] [PubMed]
- Viereck, J.; Kumarswamy, R.; Foinquinos, A.; Xiao, K.; Avramopoulos, P.; Kunz, M.; Dittrich, M.; Maetzig, T.; Zimmer, K.; Remke, J.; et al. Long noncoding RNA Chast promotes cardiac remodeling. Sci. Transl. Med. 2016, 8. [Google Scholar] [CrossRef] [PubMed]
- Scherberich, J.E. Immunological and ultrastructural analysis of loss of tubular membrane-bound enzymes in patients with renal damage. Clin. Chim. Acta 1989, 185, 271–282. [Google Scholar] [CrossRef]
- Dluzen, D.F.; Noren Hooten, N.; Zhang, Y.; Kim, Y.; Glover, F.E.; Tajuddin, S.M.; Jacob, K.D.; Zonderman, A.B.; Evans, M.K. Racial differences in microRNA and gene expression in hypertensive women. Sci. Rep. 2016, 6, 35815. [Google Scholar] [CrossRef] [PubMed]

| miRNA | miRNA Expression | Species | Conditions/Treatment | Sample Size | Source | Ref. |
|---|---|---|---|---|---|---|
| RAAS | ||||||
| miR-155 | Down | Human | young HT, reporter silencing assay | n = 19–25 | Blood; HEK293T cells | [39,40] |
| miR-526, miR-578, miR-34a, miR-34c-5p, miR-449b, miR-571, miR-765 | Up | Human | SNP genotyping on miRNA binding sites in genes of RAAS that influence blood pressure | n = 1246 | Blood, HUH7/HELA cells | [41] |
| miR-483-3p | Up | Human, rats, mice | MiRNA array, reporter luciferase assay | - | HASMC, RASMC, HL-1 cells | [42] |
| miR-143/145 | Up | Mice | Shear stress on EC of Ampkα2−/− mice | - | EC | [43] |
| - | Mice | MiR-143/145 KO mice: AngII-infusion for vascular injury | Mesenteric arteries | [44] | ||
| miR-132, miR-212 | Up | Rats | AngII-infused and Endothelin | n = 3–5 | Heart, aorta, kidney | [45] |
| down | Human | AGTR1 blocker treatmt | n = 16 | Artery | [45] | |
| miR-21 | Up | human | AngII-induced cells | Cell line | [46] | |
| miR-4516 | Human | HT iSS/SS/SR | n = 3–4 | Exosomes in urine | [47] | |
| miR-361-5p, miR-362-5p | Down | Human | SSH vs. SRH | n = 6 | Whole blood | [48] |
| miR-638,181a,663, let-7c | Down | Human | qPCR on HT/NT | n = 16–22 | Renal medulla | [49] |
| miR-21,126, 196a,451 | Up | |||||
| miR-181a | Down | Mouse | Effect of RAAS on hypertension in BPH/2J mouse circadian HT | n = 7–13 | Kidneys | [50] |
| Endothelial cells | ||||||
| miR-122 | Up | Human | HT | n = 278–498 | Blood | [51] |
| miR-505 | Up | Human | HT | n = 11–19 | Plasma, HUVEC | [52,53] |
| miR-UL112, 296-5p, let-7e | Up | Microarray, qPCR | n = 67–127 | Plasma | [54] | |
| miR-155 | Up | Human | - | n = 6 | HUVEC | [55] |
| miR-221/222 | Dicer silencing by siRNA on HUVEC, hy.926 cells | [56,57,58] | ||||
| miR-146a/b | Up | Human, mice | miR 146a−/− mice exposed to inflammatory cytokines | - | HUVEC | [59] |
| miR-126 | - | Mouse / Zebrafish | miR-126−/− mice | - | Stem cells, zebrafish | [60,61,62] |
| VSMC and other cells | ||||||
| miR-21 | Up | Human | HT patients and post antihypertensive treatment | n = 95 | Peripheral blood mononuclear cells | [63] |
| miR-143, | Down | Human | Expression analysis of miRNAs involved in VSMC plasticity | n = 29–60 | Blood cells | [64,65] |
| miR-145,miR-133 | Down | Human | ||||
| miR-21, miR-1 | Up | Human | ||||
| miR-9,126 | Down | Human | HT | n = 29–60 | Blood cells | [66] |
| miR-126 | Up | Human | HUVEC | n = 6 | HUVEC | [67] |
| miR-223 | Up | Human | High density lipoprotein | HCAEC | [68] | |
| miR-34b | Down | Rats | SHR vs. Wky | n = 36 | VSMC | [69] |
| miR-29a/b/c | Up | Human | Untreated essential hypertension vs. healthy individuals | n = 30–54 | Plasma | [70] |
| miR-510 | Up | Human | HT vs. NT | n = 208–220 | Blood | [71] |
| let-7 | Up | Human | Expression of let-7 in HT vs. NT with normal/increased CMIT | n = 60 | Plasma | [72] |
| miR-92a | Up | Human | Expression of miR-92a in HT vs. NT with normal/increased CMIT | n = 60 | Plasma | [73] |
| SNPs | ncRNA | Gene | SNP site | Ref. |
|---|---|---|---|---|
| rs3749585 | miR-495 | CORIN | miR-495 site | [78] |
| rs10757274, rs2383207, rs10757278, rs1333049 | CDKN2B-AS1 (lncRNA) | - | 9p21.3 | [81] |
| rs4705342 | - | - | miR-143 promoter | [79] |
| rs17228616 | - | ACHE | miR-608 | [77] |
| rs5068 | - | NPPA | miR-425 site | [76] |
| rs938671 | - | ATP6V0A1 | miR-637 site | [75] |
| rs5186 (A1166C) | miR-155 | AGTR1 | miR-155 site | [39,40] |
| rs11174811 | miR-526, miR-578 | AVPR1A | miR-536, miR-578 sites | [41] |
| rs5225, rs2069591 | miR-34a, miR-34c-5p, miR-449b | BDKRN2 | miR-34a, miR-34c-5p, miR-449b sites | |
| rs13306046 | miR-571, miR-765 | TBXA2R | miR-571, miR-765 sites | |
| ss52051869 | miR-122 | SLC7A1 | miR-122 site | [51] |
| Species | miRNA | SNPs/Target Gene | Subject/Model | Ref. |
|---|---|---|---|---|
| Human | miR-155 | AGTR1: rs5186 (A1166C) | qPCR on blood mononuclear cells from 64 HT (AA: 25; AC: 20; CC: 19); HUVEC cells | [39,55] |
| - | Reporter silencing assay on HEK293T | [40] | ||
| Human | miR-638, -181a, -663, let-7c | - | Microarray. Validated by qPCR. Functional studies with HEK293 cells. qPCR HT vs. NT | [49,64,65] |
| Human | miR-21, -126, -196a, -451 | - | ||
| Human | miR-145,133 | TGF-β | qPCR HT vs. NT | [64,65] |
| Human | miR-122 | SLC7A1: ss52051869 | Genotyping, sequencing, in vitro on HT | [51] |
| Human | miR-505 | FGF18 | qPCR HT vs. NT from plasma, luciferase reporter assay | [52,53] |
| Human | miR-UL112,296-5p,let-7e | Microarray and validated by qPCR on HT vs. NT | [54] | |
| Human | let-7 | - | qPCR on let-7 in HT vs. NT with normal/increased CMIT | [72] |
| Human | miR-155 | eNOS | qPCR and in vitro assay on HUVEC | [55] |
| Human | miR-143 | - | qPCR HT vs. NT | [64] |
| Human | miR-9,126 | VCAM-1, ICAM-1 | qPCR HT vs. NT | [66] |
| Human | miR-126 | VCAM | Microarray, northern blot and fucntional assay on HUVEC | [67] |
| Human | miR-223 | ICAM-1 | Whole genome and miRNA microarray on HDL treated HCAEC, qPCR, luciferase reporter assay | [68] |
| Human | miR-361-5p, miR-362-5p | - | qPCR on SSH vs. SRH | [48] |
| Human | miR-21 | - | 1.HT patients and post antihypertensive treatment. 2 AngII-induced H295R cells | [63] |
| AngII-induced H295R cells and luciferase reporter assay | [46] | |||
| Human | miR-29a/b/c | - | untreated essential hypertension vs. healthy individuals | [70] |
| Human | miR-510 | - | qPCR on HT vs. NT | [71] |
| Human | miR-92a | - | qPCR on miR-92a in HT vs. NT with normal/increased CMIT | [73] |
| Human | miR-4516 | qPCR from exosomes of urine of HT ISS/SS/SR | [47] | |
| Human | miR-221/222 | eNOS, STAT5a, Ets1, Ets2, p21Cip1, p27Kip1 | Mcroarray, Northern blotting on Dicer silenced HUVEC and and EA.hy.926 cells | [56,58] |
| Human, rats, mice | miR-132, 212 | - | Microarray. Validated by qPCR. Humans treated: AngII blocker, β-blocker; rats treated with endothelin, mice treated with AngII | [45] |
| Human, rats, mice | miR-483-3p | AT2R, AGT, ACE1, ACE2 | miRNA array, luciferase reporter assay on HASMC, RASMC, HL-1 cells | [42] |
| Human, mice | miR-146a/b | HuR | qPCR and intro assay on HUVEC and mice tissues induced by inflammatory cytokines | [59] |
| Rats | miR-34b | Cdk6 | qPCR on SHR vs. Wky | [69] |
| Rats | miR-22 | Chga | Luciferase reporter assay, miR-22 antagomir | [91] |
| Mice | miR-143/145 | ACE | Shear stress on EC of Ampkα2−/− mice, qPCR. MiRagen Therapeutics: MGN-2677 | [31,43] |
| Mouse | miR-181a | - | qPCR on BHP/2J mouse circadian HT | [50] |
| Mouse/Zebrafish | miR-126 | VCAM1, SPRED-1, PIK3 regulatory subunit-2 | miR-126−/− mice, mouse ES cells, antisense to miR-126 | [60,61] |
| Species | lncRNA | Cohort/Model | Function | Detection/Evaluation | Outcome | Ref. |
|---|---|---|---|---|---|---|
| Human | CDKN2B-AS1 | HT vs. NT (Turkish) | Interacts with PRC1 & PRC2 to repress CDKN2A/B locus. Regulate VSMC stiffness | qPCR to test if published 9p21.3 SNPs are associated with BP | Significant difference in genotype freq of the 4 SNPs betw HT and NT. Association betw rs10757274 & rs2383207 (AA) and SBP. | [81] |
| Human | H19 | 87,736 indiv. + 68,368 indiv. from European ancestry | Regulator of mammlain development, inhibits cell proliferation. Methylation of H19 associated with preeclampsia and imprinting syndrome and growth retardation. | Discovery meta analysis, genome-wide SNP genotype | 11 Loci with 31 genes uncovered with H19 as a lncRNA. | [96] |
| Human/Rat | GAS5 | Transfecton of HUVEC, human VSMC, GAS5 viral knockdown in SHR vs. Wky | Regulate remodelling of arteries (caudal, carotid, renal and thoracic); regulate transcription of androgen, progesterone, mineralcorticoid receptors; involved in cellular growth arrest and apoptosis | BP measurement, tissue staining for arterial remodeling evaluation, qPCR for GAS5 expression | GAS5 expression down regulated in HT. knockdown increased SBP and DBP and mean arterial BP (in SHR) retinal neovascularization and capillary leakage, endothelial activation and proliferation | [99] |
| Human/Rat | AK098656 | HT vs. NT (China); AK098656 transgenic rat model | Induce VSMC synthetic phenotye. Bind to myosin heavy chain-11, fibronectin-1, 26S proteasome non-ATPase regulatory subunit 11, actin, actin-binding protein | LncRNA microarray, whole-genome microarray | Upregulated in plasma of HT group vs. NT, increase VSMC proliferation & migration, upregulate extracellular matrix but downregulate contractile proteins. | [95] |
| Human/Mouse | MALAT1 | HUVEC and MALAT1 KO model | Control cellular proliferation through histone modification | RNASeq, Microarray, qPCR | Vessel growth, endothelial cell function | [97] |
| Rats | XR007793 | Wky/SHR and VSMC subjected to hypertensive level cyclic strain | No known predicted target | Microarray and qPCR | Kncockdown of XR007793 repress VSMC proliferation & migration. Reduced transcript expression of stat2, lmo2 and irf7. | [93] |
| Rats | 749 lncRNAs | Dahl SS/SR and SHR | - | RNASeq, mRNA trasncrptome analysis | Asb3, Chac2, Pex11b, Sp5 | [94] |
| Rats | sONE | Borderline hypertensive rats (BHR) fed high, medium and low salt diets | From transcription unit (NOS3AS) on opposing strand of human eNOS. Inhibiton of sONE increases eNOS and vice versa when sONE is overexpressed. | qPCR | Lycium Barbarum L. ameliorated hypertension, reduced sONE expression and improved eNOS expression compared to high salt diet rats. | [100] |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Leimena, C.; Qiu, H. Non-Coding RNA in the Pathogenesis, Progression and Treatment of Hypertension. Int. J. Mol. Sci. 2018, 19, 927. https://doi.org/10.3390/ijms19040927
Leimena C, Qiu H. Non-Coding RNA in the Pathogenesis, Progression and Treatment of Hypertension. International Journal of Molecular Sciences. 2018; 19(4):927. https://doi.org/10.3390/ijms19040927
Chicago/Turabian StyleLeimena, Christiana, and Hongyu Qiu. 2018. "Non-Coding RNA in the Pathogenesis, Progression and Treatment of Hypertension" International Journal of Molecular Sciences 19, no. 4: 927. https://doi.org/10.3390/ijms19040927
APA StyleLeimena, C., & Qiu, H. (2018). Non-Coding RNA in the Pathogenesis, Progression and Treatment of Hypertension. International Journal of Molecular Sciences, 19(4), 927. https://doi.org/10.3390/ijms19040927